Chlorine »
PDB 6ggo-6gos »
6gmu »
Chlorine in PDB 6gmu: Serum Paraoxonase-1 By Directed Evolution with the L69G/H134R/F222S/T332S Mutations
Protein crystallography data
The structure of Serum Paraoxonase-1 By Directed Evolution with the L69G/H134R/F222S/T332S Mutations, PDB code: 6gmu
was solved by
M.Ben-David,
J.L.Sussman,
D.S.Tawfik,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 33.31 / 2.70 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 94.318, 94.318, 142.410, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.9 / 22.9 |
Other elements in 6gmu:
The structure of Serum Paraoxonase-1 By Directed Evolution with the L69G/H134R/F222S/T332S Mutations also contains other interesting chemical elements:
| Calcium | (Ca) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Serum Paraoxonase-1 By Directed Evolution with the L69G/H134R/F222S/T332S Mutations
(pdb code 6gmu). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 3 binding sites of Chlorine where determined in the Serum Paraoxonase-1 By Directed Evolution with the L69G/H134R/F222S/T332S Mutations, PDB code: 6gmu:
Jump to Chlorine binding site number: 1; 2; 3;
In total 3 binding sites of Chlorine where determined in the Serum Paraoxonase-1 By Directed Evolution with the L69G/H134R/F222S/T332S Mutations, PDB code: 6gmu:
Jump to Chlorine binding site number: 1; 2; 3;
Chlorine binding site 1 out of 3 in 6gmu
Go back to
Chlorine binding site 1 out
of 3 in the Serum Paraoxonase-1 By Directed Evolution with the L69G/H134R/F222S/T332S Mutations

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Serum Paraoxonase-1 By Directed Evolution with the L69G/H134R/F222S/T332S Mutations within 5.0Å range:
|
Chlorine binding site 2 out of 3 in 6gmu
Go back to
Chlorine binding site 2 out
of 3 in the Serum Paraoxonase-1 By Directed Evolution with the L69G/H134R/F222S/T332S Mutations
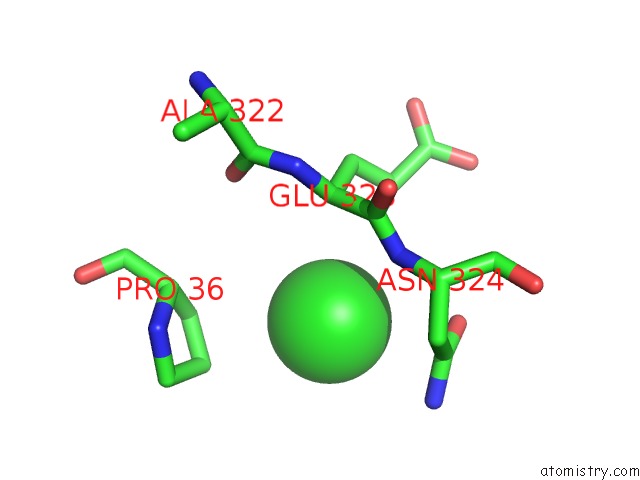
Mono view
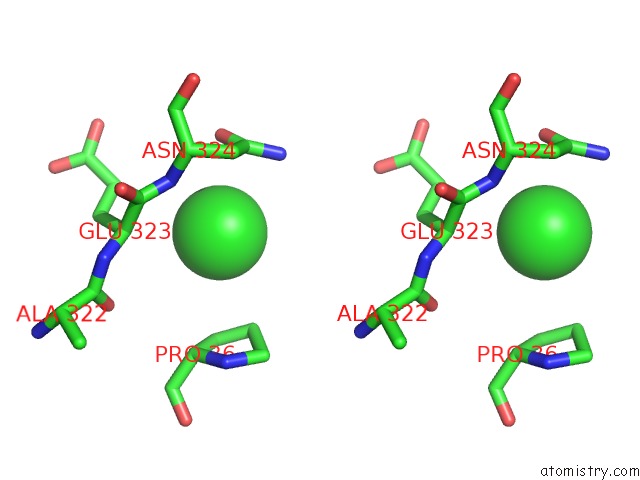
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Serum Paraoxonase-1 By Directed Evolution with the L69G/H134R/F222S/T332S Mutations within 5.0Å range:
|
Chlorine binding site 3 out of 3 in 6gmu
Go back to
Chlorine binding site 3 out
of 3 in the Serum Paraoxonase-1 By Directed Evolution with the L69G/H134R/F222S/T332S Mutations
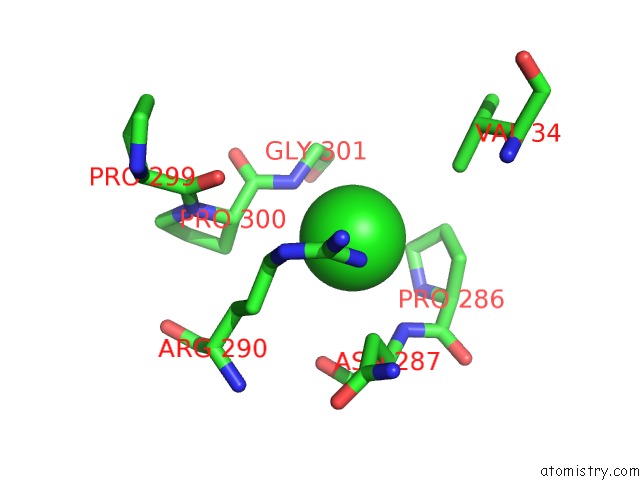
Mono view
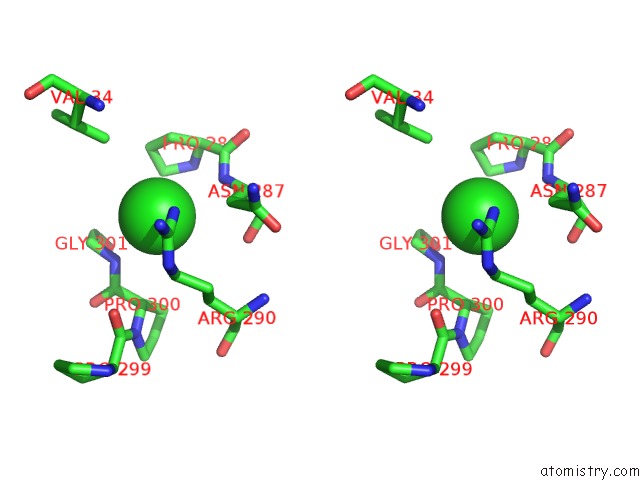
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Serum Paraoxonase-1 By Directed Evolution with the L69G/H134R/F222S/T332S Mutations within 5.0Å range:
|
Reference:
M.Ben-David,
L.S.Joel,
S.T.Dan.
An Epistatic Ratchet Versus A Reversible Transition in the Evolution of Enzyme Function To Be Published.
Page generated: Sat Jul 12 14:34:52 2025
Last articles
F in 4JLNF in 4JLT
F in 4JLM
F in 4JLG
F in 4JJU
F in 4JLK
F in 4JKV
F in 4JLJ
F in 4JJS
F in 4JII