Chlorine »
PDB 6gp4-6gxd »
6grh »
Chlorine in PDB 6grh: E. Coli Microcin Synthetase Mcbbcd Complex with Truncated Pro-MCCB17 Bound
Protein crystallography data
The structure of E. Coli Microcin Synthetase Mcbbcd Complex with Truncated Pro-MCCB17 Bound, PDB code: 6grh
was solved by
D.Ghilarov,
C.E.M.Stevenson,
D.Y.Travin,
J.Piskunova,
M.Serebryakova,
A.Maxwell,
D.M.Lawson,
K.Severinov,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 57.54 / 1.85 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 180.740, 83.760, 87.120, 90.00, 91.33, 90.00 |
| R / Rfree (%) | 17.2 / 20.7 |
Other elements in 6grh:
The structure of E. Coli Microcin Synthetase Mcbbcd Complex with Truncated Pro-MCCB17 Bound also contains other interesting chemical elements:
| Zinc | (Zn) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the E. Coli Microcin Synthetase Mcbbcd Complex with Truncated Pro-MCCB17 Bound
(pdb code 6grh). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the E. Coli Microcin Synthetase Mcbbcd Complex with Truncated Pro-MCCB17 Bound, PDB code: 6grh:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the E. Coli Microcin Synthetase Mcbbcd Complex with Truncated Pro-MCCB17 Bound, PDB code: 6grh:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 6grh
Go back to
Chlorine binding site 1 out
of 2 in the E. Coli Microcin Synthetase Mcbbcd Complex with Truncated Pro-MCCB17 Bound
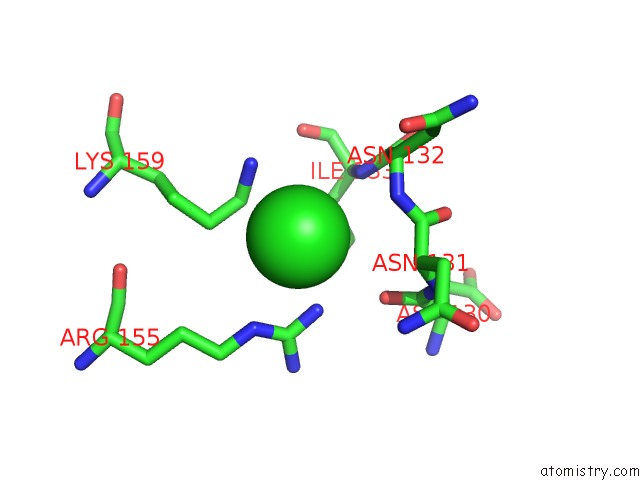
Mono view
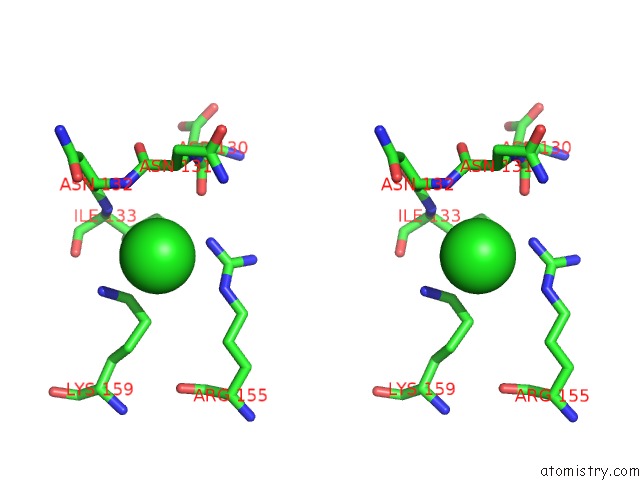
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of E. Coli Microcin Synthetase Mcbbcd Complex with Truncated Pro-MCCB17 Bound within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 6grh
Go back to
Chlorine binding site 2 out
of 2 in the E. Coli Microcin Synthetase Mcbbcd Complex with Truncated Pro-MCCB17 Bound
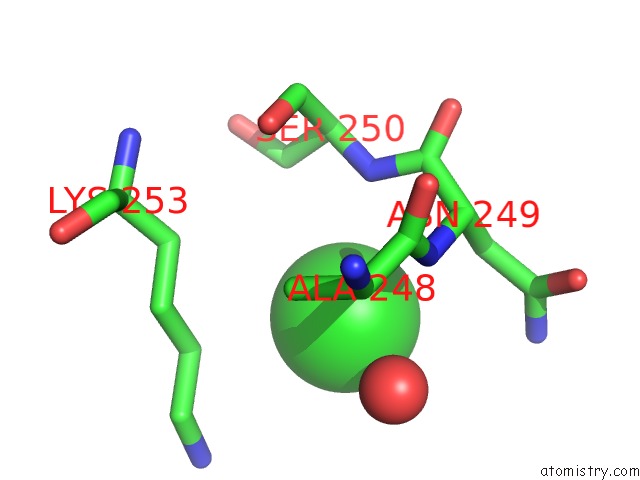
Mono view
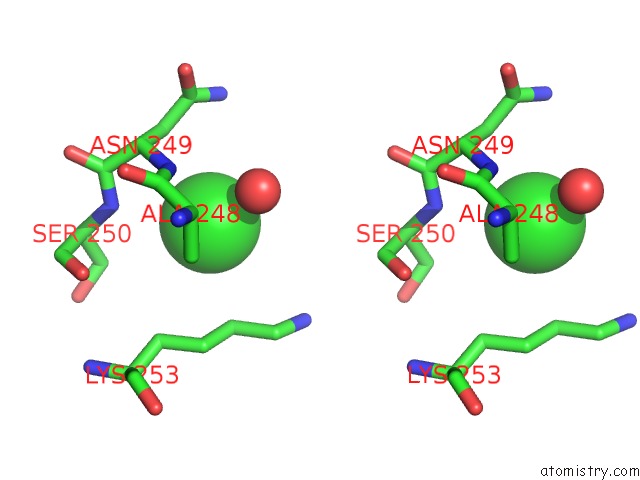
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of E. Coli Microcin Synthetase Mcbbcd Complex with Truncated Pro-MCCB17 Bound within 5.0Å range:
|
Reference:
D.Ghilarov,
C.E.M.Stevenson,
D.Y.Travin,
J.Piskunova,
M.Serebryakova,
A.Maxwell,
D.M.Lawson,
K.Severinov.
Architecture of Microcin B17 Synthetase: An Octameric Protein Complex Converting A Ribosomally Synthesized Peptide Into A Dna Gyrase Poison. Mol. Cell V. 73 749 2019.
ISSN: ISSN 1097-4164
PubMed: 30661981
DOI: 10.1016/J.MOLCEL.2018.11.032
Page generated: Sat Jul 12 14:41:00 2025
ISSN: ISSN 1097-4164
PubMed: 30661981
DOI: 10.1016/J.MOLCEL.2018.11.032
Last articles
Cl in 6QSZCl in 6QQC
Cl in 6QR1
Cl in 6QQD
Cl in 6QQN
Cl in 6QQE
Cl in 6QQF
Cl in 6QQ3
Cl in 6QPU
Cl in 6QP4