Chlorine »
PDB 6i6p-6in0 »
6iah »
Chlorine in PDB 6iah: Phosphatase TT82 From Thermococcus Thioreducens
Protein crystallography data
The structure of Phosphatase TT82 From Thermococcus Thioreducens, PDB code: 6iah
was solved by
J.Brynda,
I.K.Smatanova,
P.Pachl,
T.Prudnikova,
R.J.Masters,
P.Havlickova,
B.Kascakova,
V.Brinsa,
M.Kuty,
M.L.Pusey,
J.D.Ng,
P.M.Rezacova,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 43.87 / 1.75 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 66.330, 117.000, 33.830, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.5 / 22.6 |
Other elements in 6iah:
The structure of Phosphatase TT82 From Thermococcus Thioreducens also contains other interesting chemical elements:
| Magnesium | (Mg) | 3 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Phosphatase TT82 From Thermococcus Thioreducens
(pdb code 6iah). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 7 binding sites of Chlorine where determined in the Phosphatase TT82 From Thermococcus Thioreducens, PDB code: 6iah:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7;
In total 7 binding sites of Chlorine where determined in the Phosphatase TT82 From Thermococcus Thioreducens, PDB code: 6iah:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7;
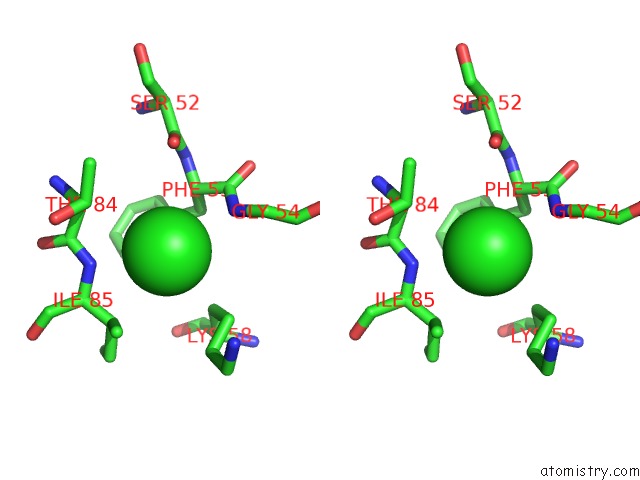
Chlorine binding site 1 out of 7 in 6iah
Go back to
Chlorine binding site 1 out
of 7 in the Phosphatase TT82 From Thermococcus Thioreducens

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Phosphatase TT82 From Thermococcus Thioreducens within 5.0Å range:
|
Chlorine binding site 2 out of 7 in 6iah
Go back to
Chlorine binding site 2 out
of 7 in the Phosphatase TT82 From Thermococcus Thioreducens

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Phosphatase TT82 From Thermococcus Thioreducens within 5.0Å range:
|
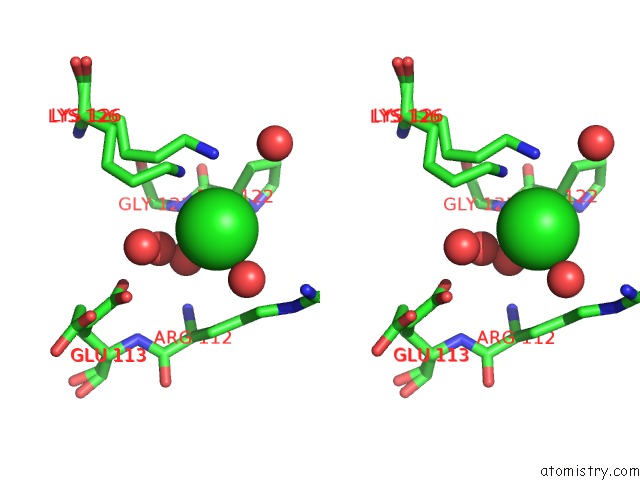
Chlorine binding site 3 out of 7 in 6iah
Go back to
Chlorine binding site 3 out
of 7 in the Phosphatase TT82 From Thermococcus Thioreducens

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Phosphatase TT82 From Thermococcus Thioreducens within 5.0Å range:
|
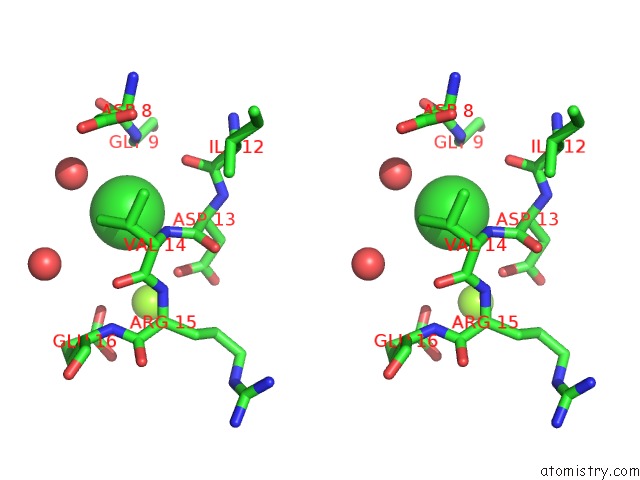
Chlorine binding site 4 out of 7 in 6iah
Go back to
Chlorine binding site 4 out
of 7 in the Phosphatase TT82 From Thermococcus Thioreducens

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Phosphatase TT82 From Thermococcus Thioreducens within 5.0Å range:
|
Chlorine binding site 5 out of 7 in 6iah
Go back to
Chlorine binding site 5 out
of 7 in the Phosphatase TT82 From Thermococcus Thioreducens

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Phosphatase TT82 From Thermococcus Thioreducens within 5.0Å range:
|
Chlorine binding site 6 out of 7 in 6iah
Go back to
Chlorine binding site 6 out
of 7 in the Phosphatase TT82 From Thermococcus Thioreducens

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Phosphatase TT82 From Thermococcus Thioreducens within 5.0Å range:
|
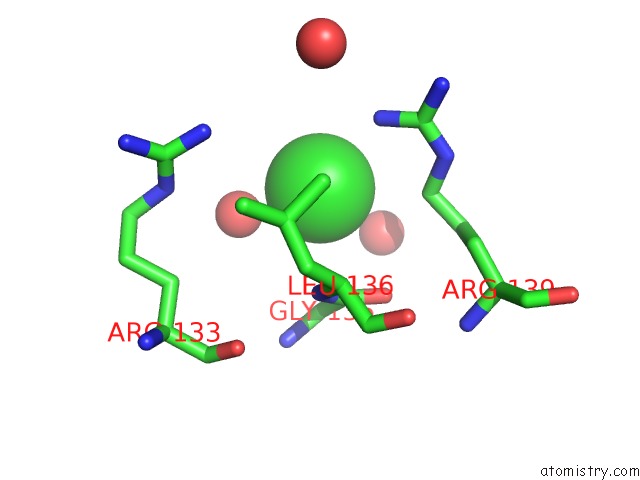
Chlorine binding site 7 out of 7 in 6iah
Go back to
Chlorine binding site 7 out
of 7 in the Phosphatase TT82 From Thermococcus Thioreducens
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 7 of Phosphatase TT82 From Thermococcus Thioreducens within 5.0Å range:
|
Reference:
P.Havlickova,
V.Brinsa,
J.Brynda,
P.Pachl,
T.Prudnikova,
J.R.Mesters,
B.Kascakova,
M.Kuty,
M.L.Pusey,
J.D.Ng,
P.Rezacova,
I.Kuta Smatanova.
A Novel Structurally Characterized Haloacid Dehalogenase Superfamily Phosphatase From Thermococcus Thioreducens with Diverse Substrate Specificity. Acta Crystallogr D Struct V. 75 743 2019BIOL.
ISSN: ISSN 2059-7983
PubMed: 31373573
DOI: 10.1107/S2059798319009586
Page generated: Sun Jul 28 01:39:14 2024
ISSN: ISSN 2059-7983
PubMed: 31373573
DOI: 10.1107/S2059798319009586
Last articles
Cl in 2W71Cl in 2W9N
Cl in 2W7Z
Cl in 2W98
Cl in 2W8H
Cl in 2W7T
Cl in 2W70
Cl in 2W79
Cl in 2W5P
Cl in 2W6Q