Chlorine »
PDB 6kkr-6kw1 »
6kku »
Chlorine in PDB 6kku: Human KCC1 Structure Determined in Nacl and Gdn
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Human KCC1 Structure Determined in Nacl and Gdn
(pdb code 6kku). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Human KCC1 Structure Determined in Nacl and Gdn, PDB code: 6kku:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Human KCC1 Structure Determined in Nacl and Gdn, PDB code: 6kku:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 6kku
Go back to
Chlorine binding site 1 out
of 2 in the Human KCC1 Structure Determined in Nacl and Gdn
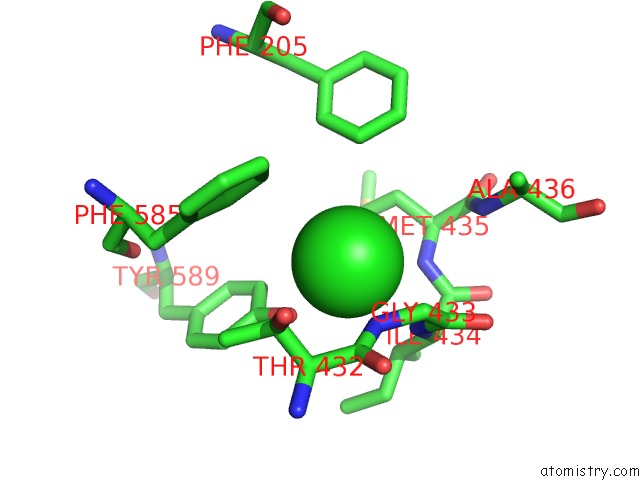
Mono view
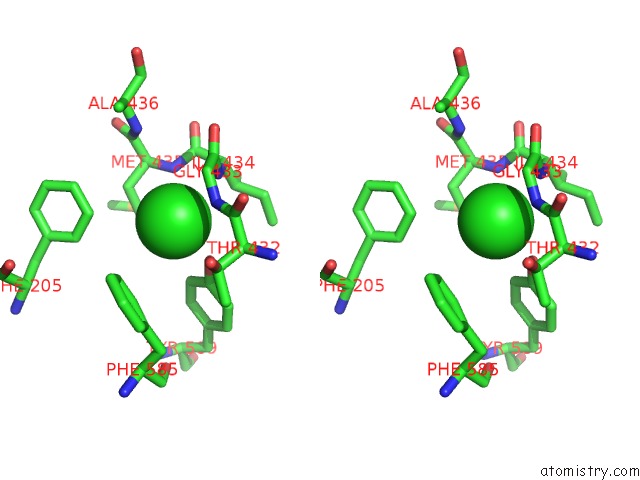
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Human KCC1 Structure Determined in Nacl and Gdn within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 6kku
Go back to
Chlorine binding site 2 out
of 2 in the Human KCC1 Structure Determined in Nacl and Gdn
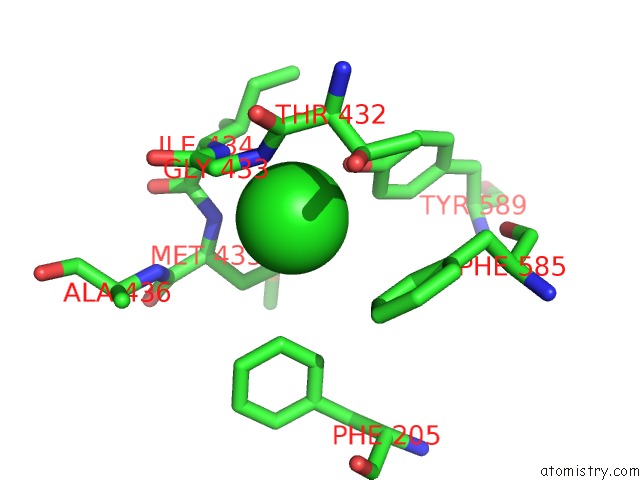
Mono view
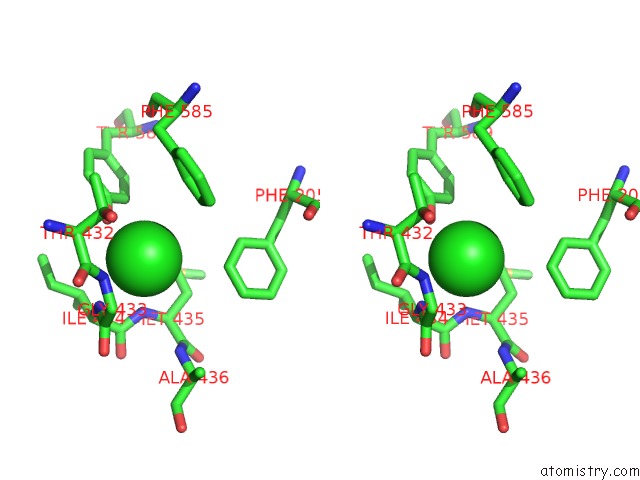
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Human KCC1 Structure Determined in Nacl and Gdn within 5.0Å range:
|
Reference:
S.Liu,
S.Chang,
B.Han,
L.Xu,
M.Zhang,
C.Zhao,
W.Yang,
F.Wang,
J.Li,
E.Delpire,
S.Ye,
X.C.Bai,
J.Guo.
Cryo-Em Structures of the Human Cation-Chloride Cotransporter KCC1. Science V. 366 505 2019.
ISSN: ESSN 1095-9203
PubMed: 31649201
DOI: 10.1126/SCIENCE.AAY3129
Page generated: Sun Jul 28 02:30:15 2024
ISSN: ESSN 1095-9203
PubMed: 31649201
DOI: 10.1126/SCIENCE.AAY3129
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1