Chlorine »
PDB 6kkr-6kw1 »
6ku2 »
Chlorine in PDB 6ku2: The Structure of Eanb/Y353A Complex with Ergothioneine Covalent Linked with Persulfide CYS412
Protein crystallography data
The structure of The Structure of Eanb/Y353A Complex with Ergothioneine Covalent Linked with Persulfide CYS412, PDB code: 6ku2
was solved by
L.Wu,
P.H.Liu,
J.H.Zhou,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.34 / 2.34 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 62.683, 88.240, 90.771, 90.00, 90.35, 90.00 |
| R / Rfree (%) | 17.1 / 22.8 |
Other elements in 6ku2:
The structure of The Structure of Eanb/Y353A Complex with Ergothioneine Covalent Linked with Persulfide CYS412 also contains other interesting chemical elements:
| Magnesium | (Mg) | 2 atoms |
| Zinc | (Zn) | 2 atoms |
| Bromine | (Br) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the The Structure of Eanb/Y353A Complex with Ergothioneine Covalent Linked with Persulfide CYS412
(pdb code 6ku2). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 3 binding sites of Chlorine where determined in the The Structure of Eanb/Y353A Complex with Ergothioneine Covalent Linked with Persulfide CYS412, PDB code: 6ku2:
Jump to Chlorine binding site number: 1; 2; 3;
In total 3 binding sites of Chlorine where determined in the The Structure of Eanb/Y353A Complex with Ergothioneine Covalent Linked with Persulfide CYS412, PDB code: 6ku2:
Jump to Chlorine binding site number: 1; 2; 3;
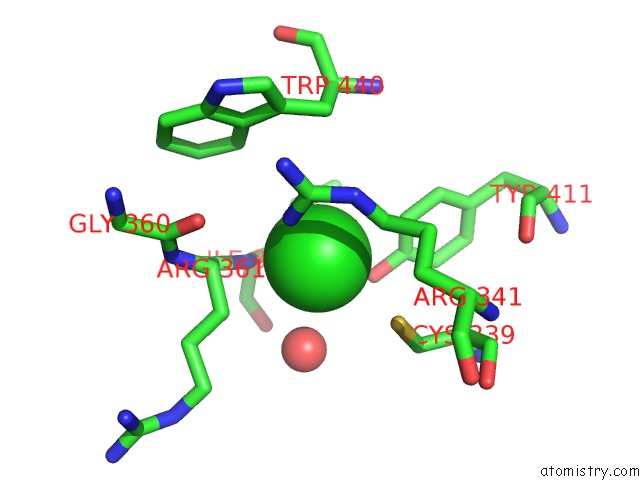
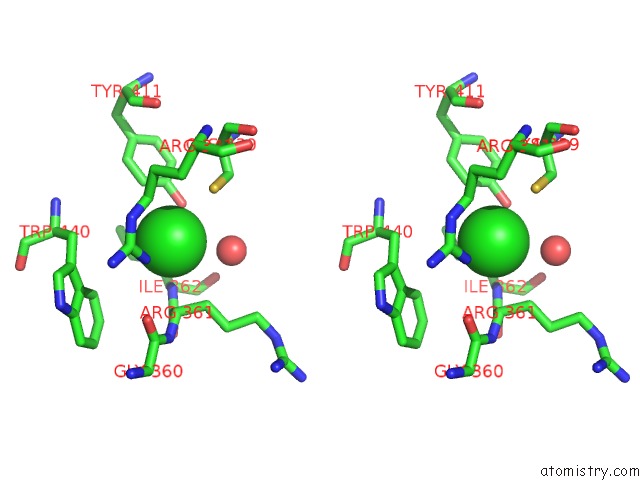
Chlorine binding site 1 out of 3 in 6ku2
Go back to
Chlorine binding site 1 out
of 3 in the The Structure of Eanb/Y353A Complex with Ergothioneine Covalent Linked with Persulfide CYS412
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of The Structure of Eanb/Y353A Complex with Ergothioneine Covalent Linked with Persulfide CYS412 within 5.0Å range:
|
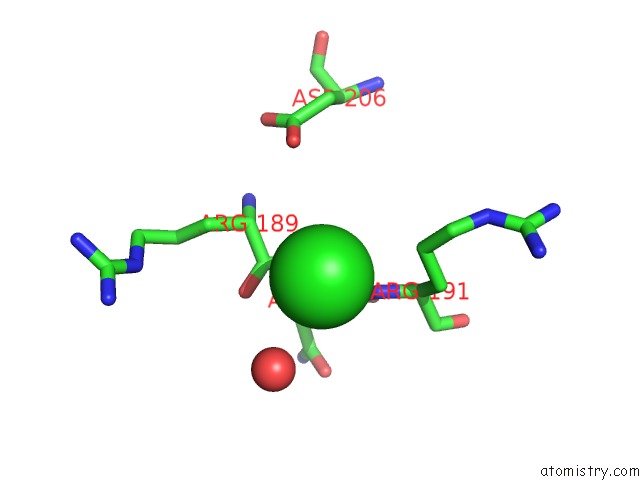
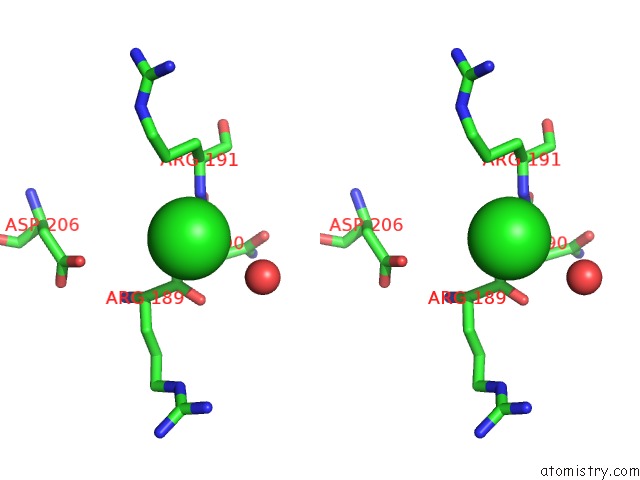
Chlorine binding site 2 out of 3 in 6ku2
Go back to
Chlorine binding site 2 out
of 3 in the The Structure of Eanb/Y353A Complex with Ergothioneine Covalent Linked with Persulfide CYS412
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of The Structure of Eanb/Y353A Complex with Ergothioneine Covalent Linked with Persulfide CYS412 within 5.0Å range:
|
Chlorine binding site 3 out of 3 in 6ku2
Go back to
Chlorine binding site 3 out
of 3 in the The Structure of Eanb/Y353A Complex with Ergothioneine Covalent Linked with Persulfide CYS412
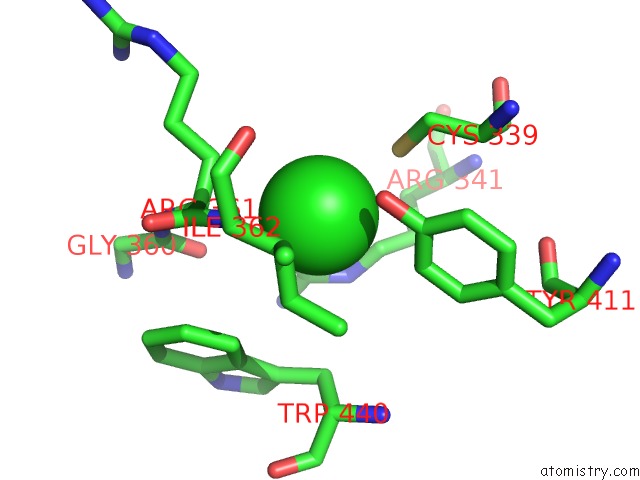
Mono view
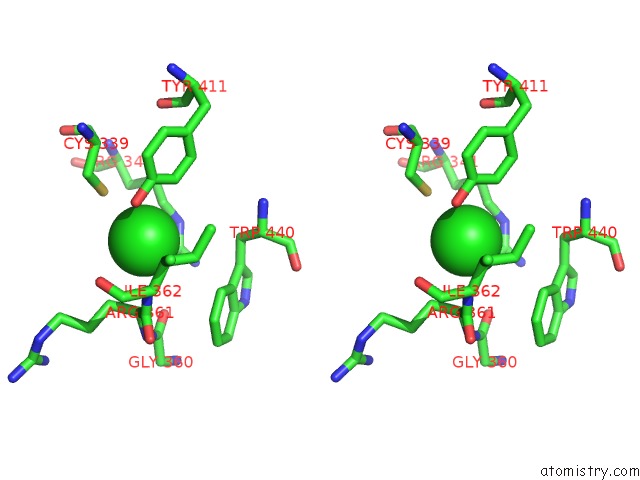
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of The Structure of Eanb/Y353A Complex with Ergothioneine Covalent Linked with Persulfide CYS412 within 5.0Å range:
|
Reference:
R.Cheng,
L.Wu,
R.Lai,
C.Peng,
N.Naowarojna,
W.Hu,
X.Li,
S.A.Whelan,
N.Lee,
J.Lopez,
C.Zhao,
Y.Yong,
J.Xue,
X.Jiang,
M.W.Grinstaff,
Z.Deng,
J.Chen,
Q.Cui,
J.H.Zhou,
P.Liu.
Single-Step Replacement of An Unreactive C-H Bond By A C-S Bond Using Polysulfide As the Direct Sulfur Source in the Anaerobic Ergothioneine Biosynthesis Acs Catalysis V. 10 8981 2020.
ISSN: ESSN 2155-5435
DOI: 10.1021/ACSCATAL.0C01809
Page generated: Sat Jul 12 16:22:53 2025
ISSN: ESSN 2155-5435
DOI: 10.1021/ACSCATAL.0C01809
Last articles
F in 7QMUF in 7QMV
F in 7QMT
F in 7QMS
F in 7QMQ
F in 7QMR
F in 7QMP
F in 7QMO
F in 7QMN
F in 7QMM