Chlorine »
PDB 6mr5-6n3v »
6n3d »
Chlorine in PDB 6n3d: Structure of Hiv Tat-Specific Factor 1 U2AF Homology Motif (Apo-State)
Protein crystallography data
The structure of Structure of Hiv Tat-Specific Factor 1 U2AF Homology Motif (Apo-State), PDB code: 6n3d
was solved by
S.Loerch,
J.L.Jenkins,
C.L.Kielkopf,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.20 / 1.13 |
| Space group | P 43 |
| Cell size a, b, c (Å), α, β, γ (°) | 30.201, 30.201, 98.742, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 12.7 / 16 |
Other elements in 6n3d:
The structure of Structure of Hiv Tat-Specific Factor 1 U2AF Homology Motif (Apo-State) also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Structure of Hiv Tat-Specific Factor 1 U2AF Homology Motif (Apo-State)
(pdb code 6n3d). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 4 binding sites of Chlorine where determined in the Structure of Hiv Tat-Specific Factor 1 U2AF Homology Motif (Apo-State), PDB code: 6n3d:
Jump to Chlorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Chlorine where determined in the Structure of Hiv Tat-Specific Factor 1 U2AF Homology Motif (Apo-State), PDB code: 6n3d:
Jump to Chlorine binding site number: 1; 2; 3; 4;
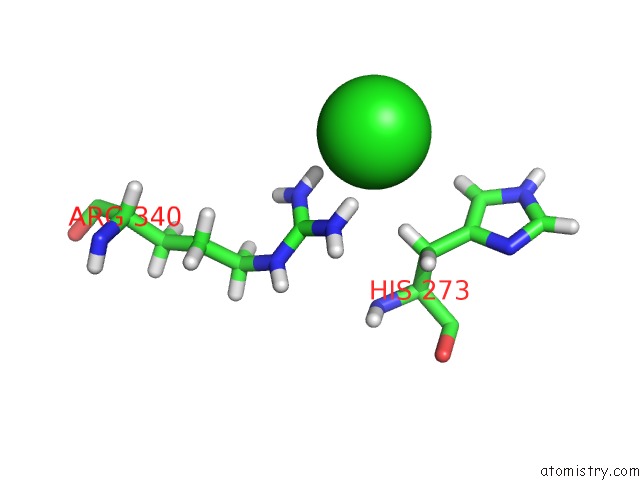
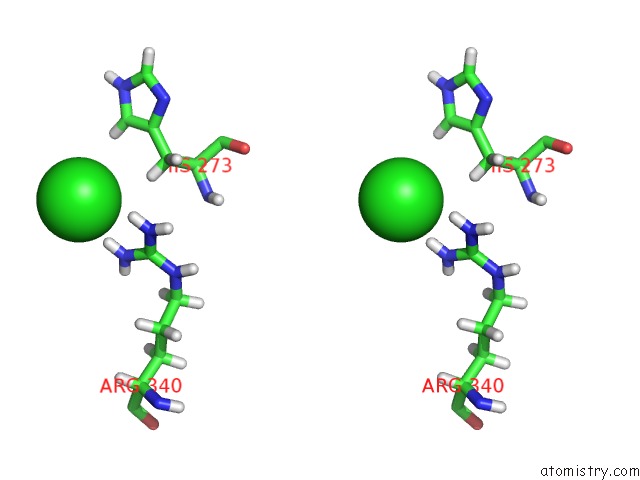
Chlorine binding site 1 out of 4 in 6n3d
Go back to
Chlorine binding site 1 out
of 4 in the Structure of Hiv Tat-Specific Factor 1 U2AF Homology Motif (Apo-State)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Structure of Hiv Tat-Specific Factor 1 U2AF Homology Motif (Apo-State) within 5.0Å range:
|
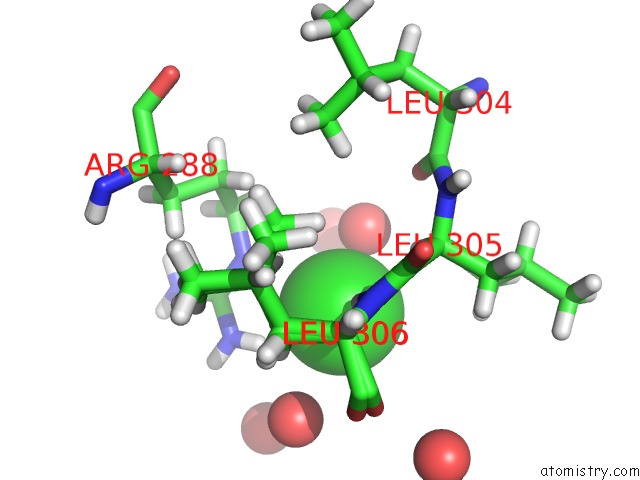
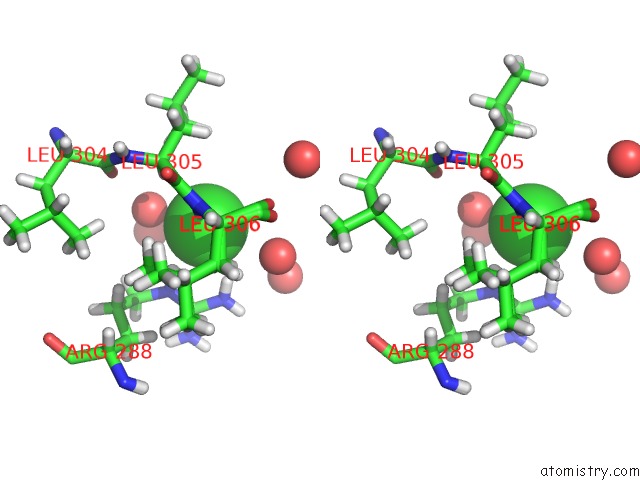
Chlorine binding site 2 out of 4 in 6n3d
Go back to
Chlorine binding site 2 out
of 4 in the Structure of Hiv Tat-Specific Factor 1 U2AF Homology Motif (Apo-State)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Structure of Hiv Tat-Specific Factor 1 U2AF Homology Motif (Apo-State) within 5.0Å range:
|
Chlorine binding site 3 out of 4 in 6n3d
Go back to
Chlorine binding site 3 out
of 4 in the Structure of Hiv Tat-Specific Factor 1 U2AF Homology Motif (Apo-State)
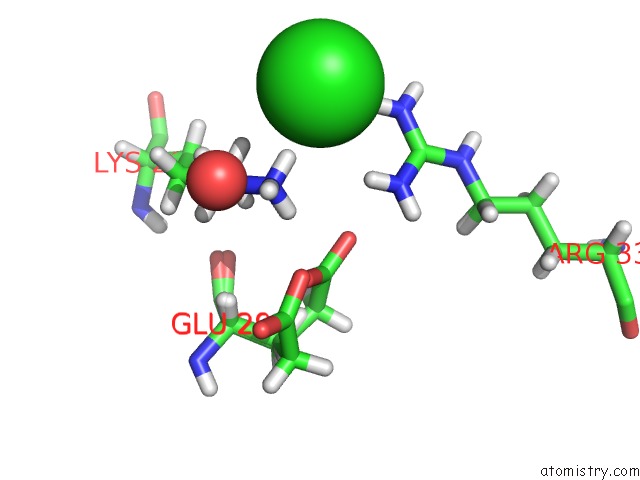
Mono view
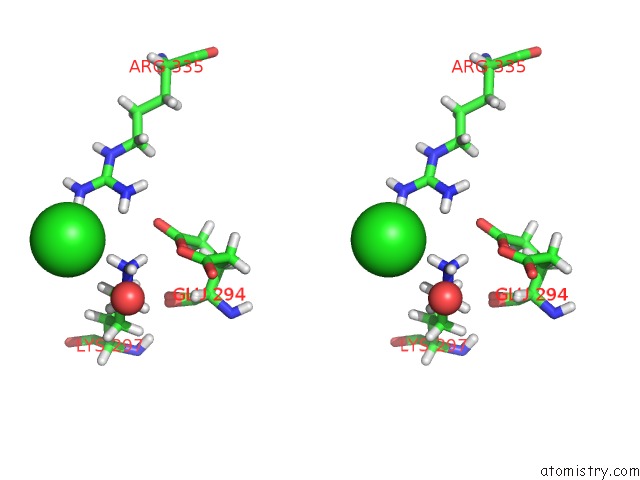
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Structure of Hiv Tat-Specific Factor 1 U2AF Homology Motif (Apo-State) within 5.0Å range:
|
Chlorine binding site 4 out of 4 in 6n3d
Go back to
Chlorine binding site 4 out
of 4 in the Structure of Hiv Tat-Specific Factor 1 U2AF Homology Motif (Apo-State)
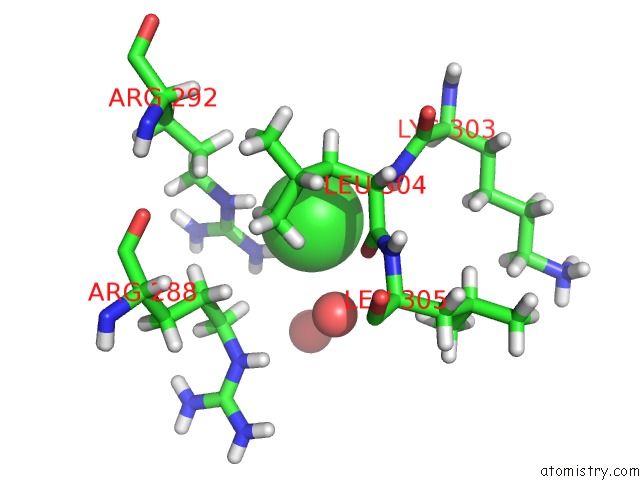
Mono view
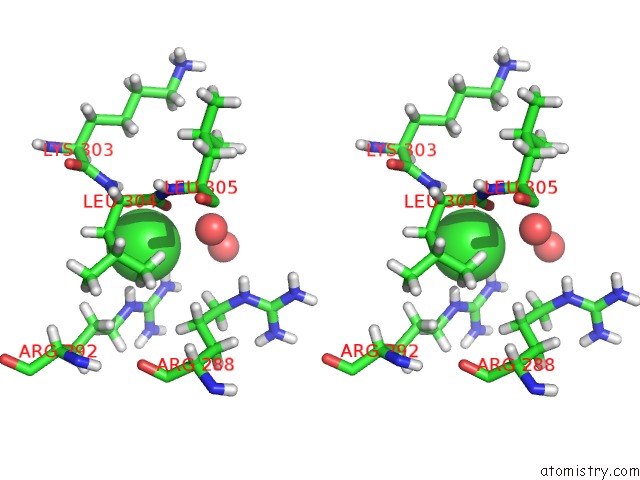
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Structure of Hiv Tat-Specific Factor 1 U2AF Homology Motif (Apo-State) within 5.0Å range:
|
Reference:
S.Loerch,
J.R.Leach,
S.W.Horner,
D.Maji,
J.L.Jenkins,
M.J.Pulvino,
C.L.Kielkopf.
The Pre-Mrna Splicing and Transcription Factor Tat-SF1 Is A Functional Partner of the Spliceosome SF3B1 Subunit Via A U2AF Homology Motif Interface. J. Biol. Chem. V. 294 2892 2019.
ISSN: ESSN 1083-351X
PubMed: 30567737
DOI: 10.1074/JBC.RA118.006764
Page generated: Sun Jul 28 03:30:14 2024
ISSN: ESSN 1083-351X
PubMed: 30567737
DOI: 10.1074/JBC.RA118.006764
Last articles
Ca in 5NMRCa in 5NN9
Ca in 5NM8
Ca in 5NH8
Ca in 5NL7
Ca in 5NIN
Ca in 5NGQ
Ca in 5NH5
Ca in 5NGY
Ca in 5NG1