Chlorine »
PDB 6nvh-6o0v »
6nzz »
Chlorine in PDB 6nzz: LRRC8A-Dcpib in MSP1E3D1 Nanodisc Expanded State
Chlorine Binding Sites:
The binding sites of Chlorine atom in the LRRC8A-Dcpib in MSP1E3D1 Nanodisc Expanded State
(pdb code 6nzz). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the LRRC8A-Dcpib in MSP1E3D1 Nanodisc Expanded State, PDB code: 6nzz:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the LRRC8A-Dcpib in MSP1E3D1 Nanodisc Expanded State, PDB code: 6nzz:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 6nzz
Go back to
Chlorine binding site 1 out
of 2 in the LRRC8A-Dcpib in MSP1E3D1 Nanodisc Expanded State
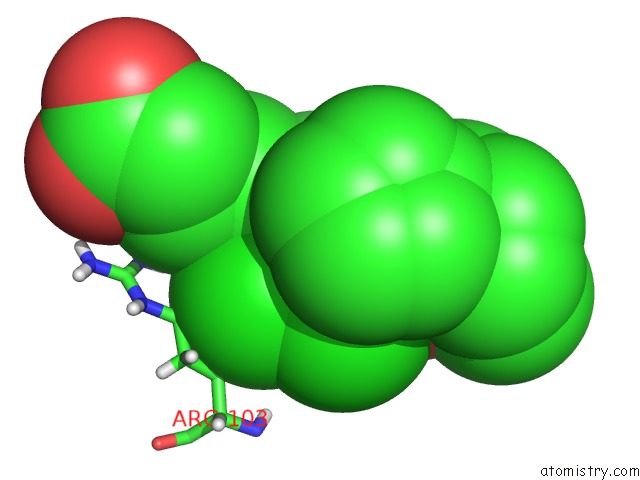
Mono view
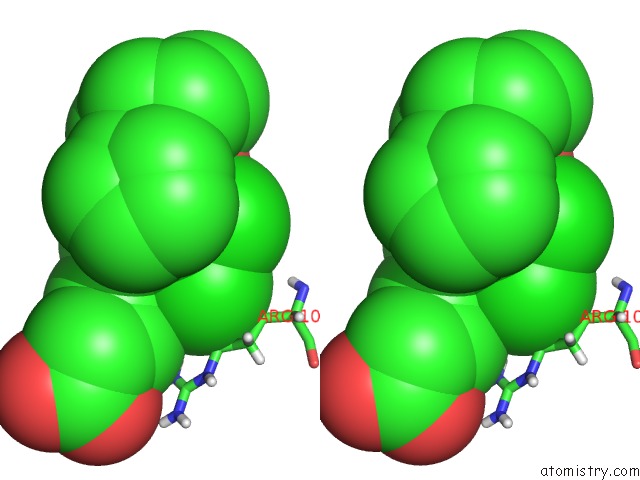
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of LRRC8A-Dcpib in MSP1E3D1 Nanodisc Expanded State within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 6nzz
Go back to
Chlorine binding site 2 out
of 2 in the LRRC8A-Dcpib in MSP1E3D1 Nanodisc Expanded State
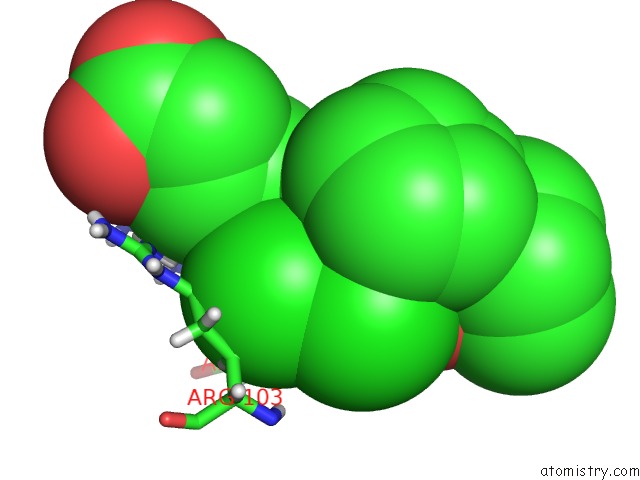
Mono view
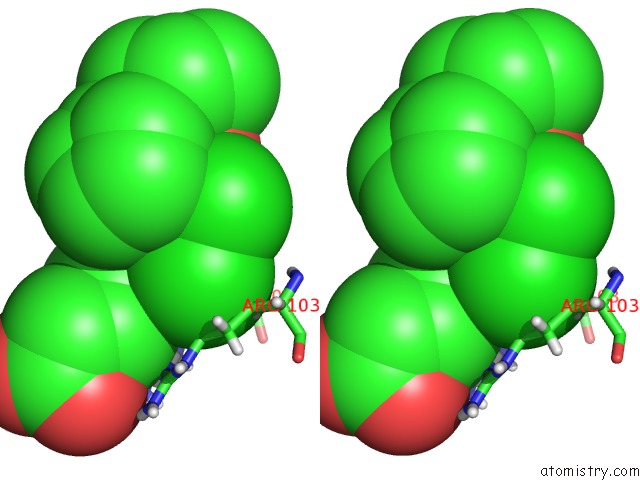
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of LRRC8A-Dcpib in MSP1E3D1 Nanodisc Expanded State within 5.0Å range:
|
Reference:
D.M.Kern,
S.Oh,
R.K.Hite,
S.G.Brohawn.
Cryo-Em Structures of the Dcpib-Inhibited Volume-Regulated Anion Channel LRRC8A in Lipid Nanodiscs. Elife V. 8 2019.
ISSN: ESSN 2050-084X
PubMed: 30775971
DOI: 10.7554/ELIFE.42636
Page generated: Mon Jul 29 12:22:09 2024
ISSN: ESSN 2050-084X
PubMed: 30775971
DOI: 10.7554/ELIFE.42636
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1