Chlorine »
PDB 6ris-6ro5 »
6rlc »
Chlorine in PDB 6rlc: Crystal Structure of the Pdz Tandem of Syntenin in Complex with Fragment F13
Protein crystallography data
The structure of Crystal Structure of the Pdz Tandem of Syntenin in Complex with Fragment F13, PDB code: 6rlc
was solved by
M.Feracci,
K.Barral,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.02 / 2.20 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 29.23, 57.512, 101.094, 93.99, 96.08, 107.7 |
| R / Rfree (%) | 25.9 / 30.1 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of the Pdz Tandem of Syntenin in Complex with Fragment F13
(pdb code 6rlc). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 4 binding sites of Chlorine where determined in the Crystal Structure of the Pdz Tandem of Syntenin in Complex with Fragment F13, PDB code: 6rlc:
Jump to Chlorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Chlorine where determined in the Crystal Structure of the Pdz Tandem of Syntenin in Complex with Fragment F13, PDB code: 6rlc:
Jump to Chlorine binding site number: 1; 2; 3; 4;
Chlorine binding site 1 out of 4 in 6rlc
Go back to
Chlorine binding site 1 out
of 4 in the Crystal Structure of the Pdz Tandem of Syntenin in Complex with Fragment F13
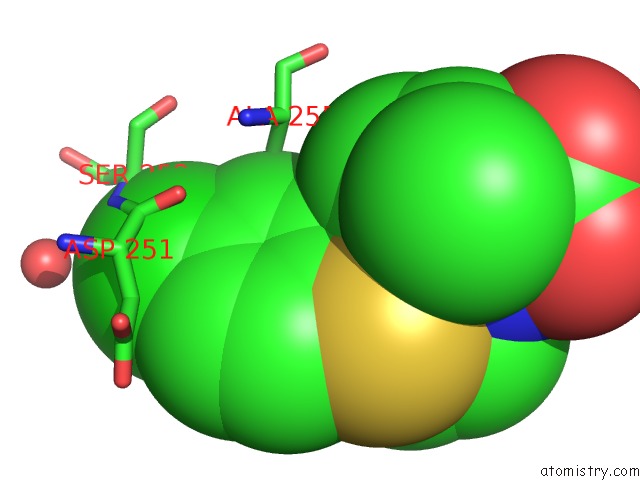
Mono view
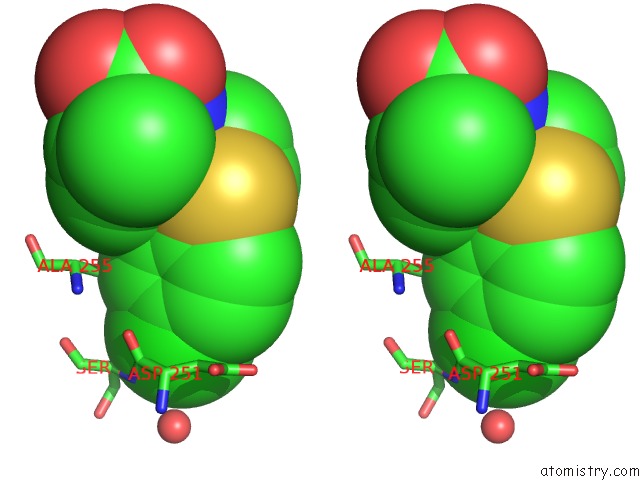
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of the Pdz Tandem of Syntenin in Complex with Fragment F13 within 5.0Å range:
|
Chlorine binding site 2 out of 4 in 6rlc
Go back to
Chlorine binding site 2 out
of 4 in the Crystal Structure of the Pdz Tandem of Syntenin in Complex with Fragment F13
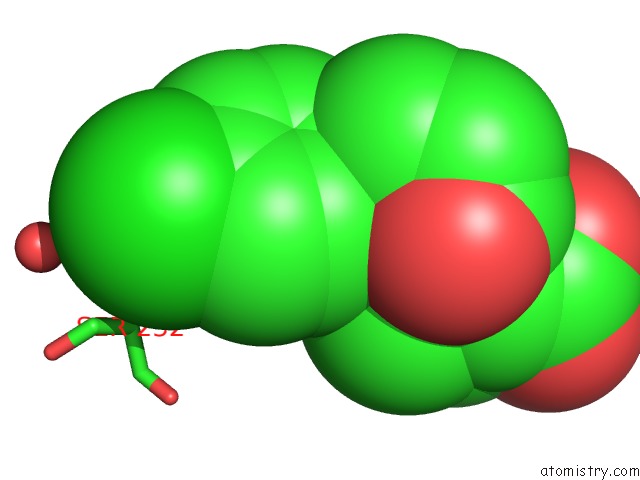
Mono view
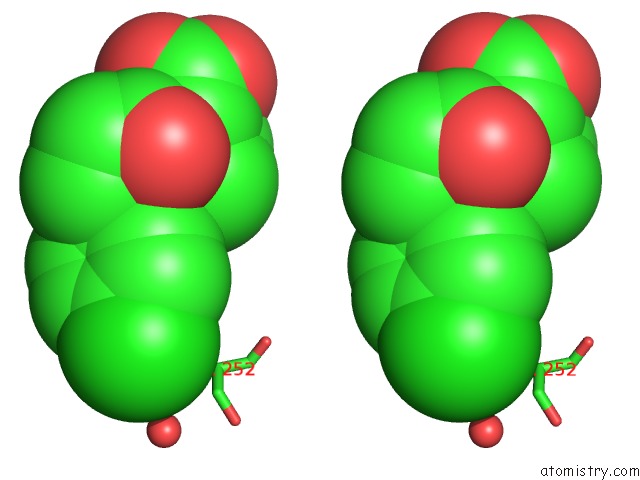
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of the Pdz Tandem of Syntenin in Complex with Fragment F13 within 5.0Å range:
|
Chlorine binding site 3 out of 4 in 6rlc
Go back to
Chlorine binding site 3 out
of 4 in the Crystal Structure of the Pdz Tandem of Syntenin in Complex with Fragment F13
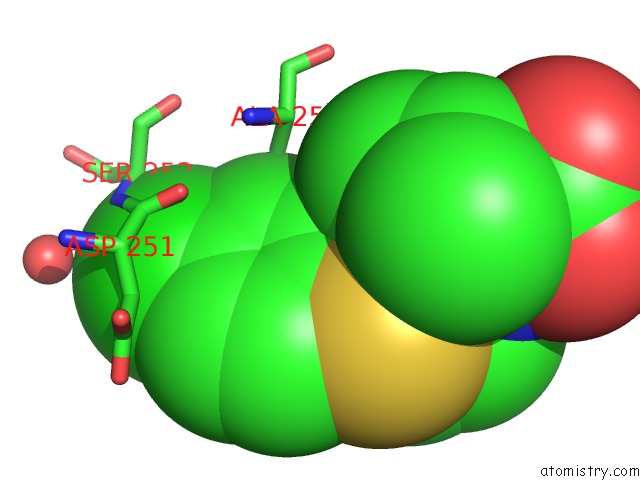
Mono view
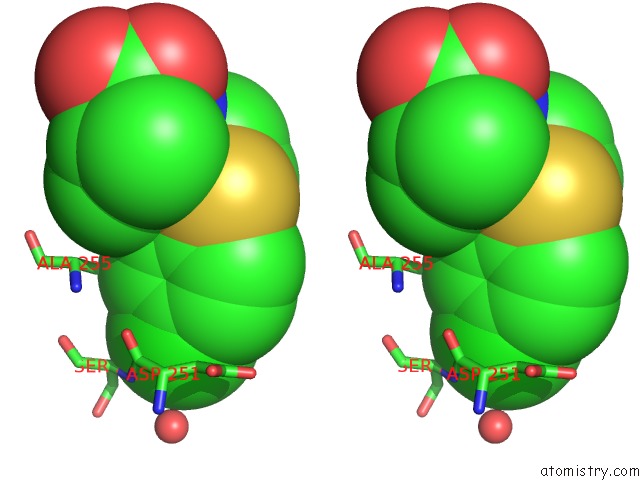
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of the Pdz Tandem of Syntenin in Complex with Fragment F13 within 5.0Å range:
|
Chlorine binding site 4 out of 4 in 6rlc
Go back to
Chlorine binding site 4 out
of 4 in the Crystal Structure of the Pdz Tandem of Syntenin in Complex with Fragment F13
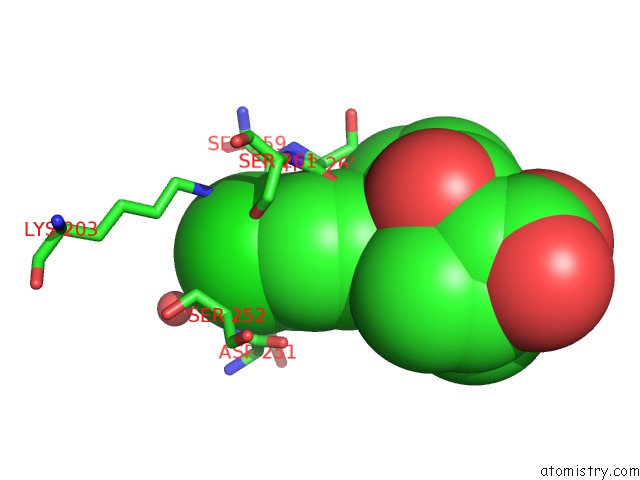
Mono view
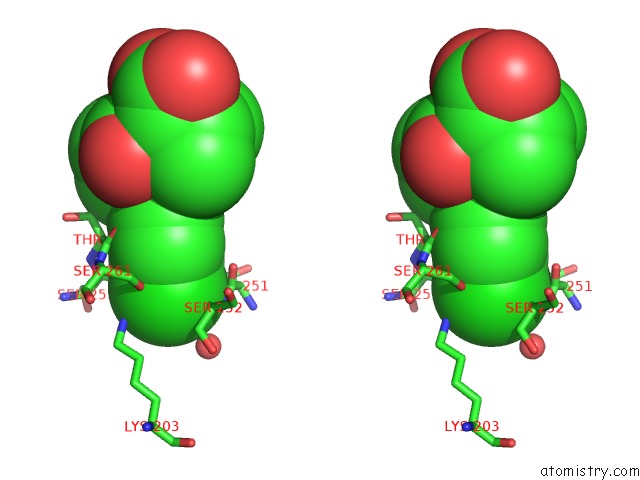
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of the Pdz Tandem of Syntenin in Complex with Fragment F13 within 5.0Å range:
|
Reference:
R.Leblanc,
R.Kashyap,
K.Barral,
A.L.Egea-Jimenez,
D.Kovalskyy,
M.Feracci,
M.Garcia,
C.Derviaux,
S.Betzi,
R.Ghossoub,
M.Platonov,
P.Roche,
X.Morelli,
L.Hoffer,
P.Zimmermann.
Pharmacological Inhibition of Syntenin PDZ2 Domain Impairs Breast Cancer Cell Activities and Exosome Loadifing with Syndecan and Epcam Cargo. J Extracell Vesicles V. 10 12039 2020.
ISSN: ISSN 2001-3078
PubMed: 33343836
DOI: 10.1002/JEV2.12039
Page generated: Mon Jul 29 14:33:27 2024
ISSN: ISSN 2001-3078
PubMed: 33343836
DOI: 10.1002/JEV2.12039
Last articles
Cl in 2XK6Cl in 2XK3
Cl in 2XJZ
Cl in 2XJW
Cl in 2XJV
Cl in 2XJU
Cl in 2XJT
Cl in 2XJO
Cl in 2XJS
Cl in 2XJR