Chlorine »
PDB 6yfw-6ynm »
6ym4 »
Chlorine in PDB 6ym4: Crystal Structure of Bay-297 with PIP4K2A
Enzymatic activity of Crystal Structure of Bay-297 with PIP4K2A
All present enzymatic activity of Crystal Structure of Bay-297 with PIP4K2A:
2.7.1.149;
2.7.1.149;
Protein crystallography data
The structure of Crystal Structure of Bay-297 with PIP4K2A, PDB code: 6ym4
was solved by
S.J.Holton,
L.Wortmann,
N.Braeuer,
H.Irlbacher,
J.Weiske,
C.Lechner,
R.Meier,
V.Puetter,
C.Christ,
T.Ter Laak,
P.Lienau,
R.Lesche,
B.Nicke,
M.Bauser,
A.Haegebarth,
F.Von Nussbaum,
D.Mumberg,
C.Lemos,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 46.07 / 1.95 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 43.984, 99.26, 104.191, 90, 93.37, 90 |
| R / Rfree (%) | 20 / 24.8 |
Other elements in 6ym4:
The structure of Crystal Structure of Bay-297 with PIP4K2A also contains other interesting chemical elements:
| Fluorine | (F) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Bay-297 with PIP4K2A
(pdb code 6ym4). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Crystal Structure of Bay-297 with PIP4K2A, PDB code: 6ym4:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Crystal Structure of Bay-297 with PIP4K2A, PDB code: 6ym4:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 6ym4
Go back to
Chlorine binding site 1 out
of 2 in the Crystal Structure of Bay-297 with PIP4K2A
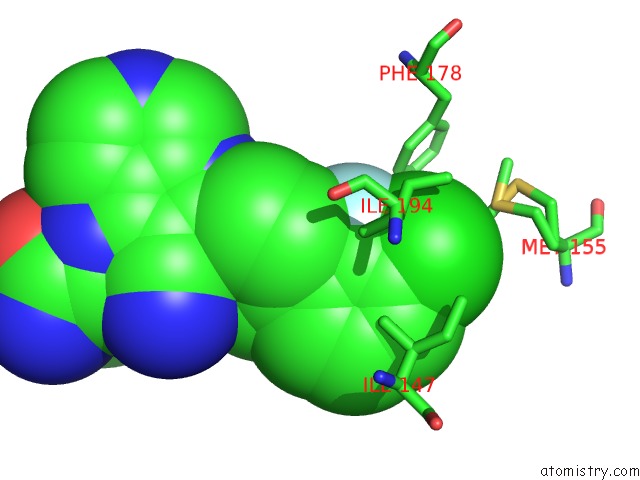
Mono view
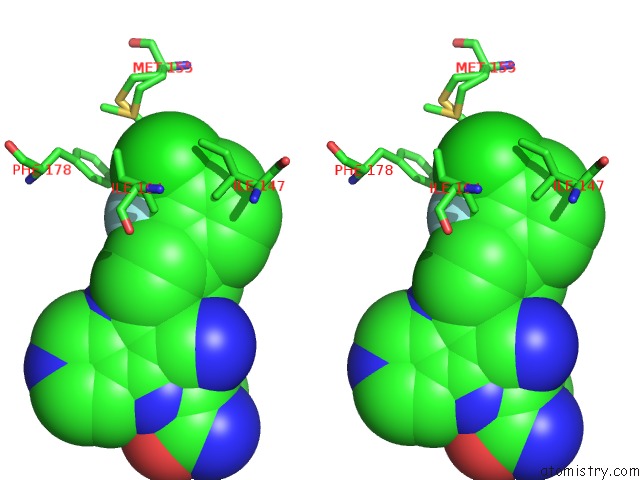
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Bay-297 with PIP4K2A within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 6ym4
Go back to
Chlorine binding site 2 out
of 2 in the Crystal Structure of Bay-297 with PIP4K2A
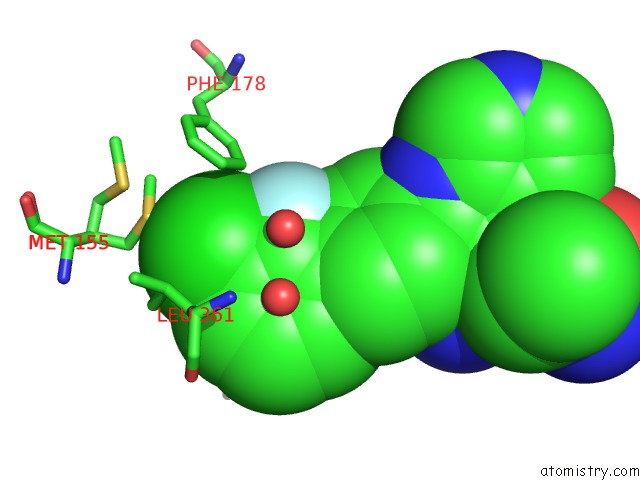
Mono view
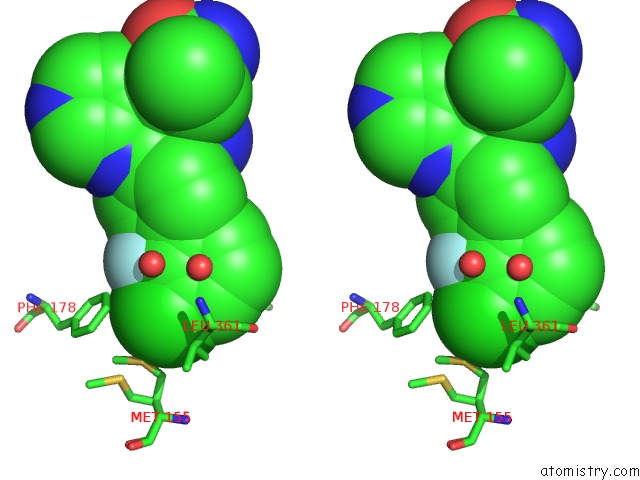
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Bay-297 with PIP4K2A within 5.0Å range:
|
Reference:
S.J.Holton,
L.Wortmann,
N.Braeuer,
H.Irlbacher,
J.Weiske,
C.Lechner,
R.Meier,
V.Puetter,
C.Christ,
T.Ter Laak,
P.Lienau,
R.Lesche,
B.Nicke,
M.Bauser,
A.Haegebarth,
F.Von Nussbaum,
D.Mumberg,
C.Lemos.
Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine Based Inhibitors Bay-091 and Bay-297 For the Kinase PIP4K2A To Be Published.
Page generated: Mon Jul 29 17:48:17 2024
Last articles
Cl in 2WIUCl in 2WJP
Cl in 2WJ2
Cl in 2WJ9
Cl in 2WJ1
Cl in 2WIK
Cl in 2WIL
Cl in 2WIG
Cl in 2WIJ
Cl in 2WID