Chlorine »
PDB 6yyf-6zc8 »
6z2v »
Chlorine in PDB 6z2v: CLK3 A319V Mutant Bound with Beta-Carboline Kh-CARB13 (Cpd 3)
Enzymatic activity of CLK3 A319V Mutant Bound with Beta-Carboline Kh-CARB13 (Cpd 3)
All present enzymatic activity of CLK3 A319V Mutant Bound with Beta-Carboline Kh-CARB13 (Cpd 3):
2.7.12.1;
2.7.12.1;
Protein crystallography data
The structure of CLK3 A319V Mutant Bound with Beta-Carboline Kh-CARB13 (Cpd 3), PDB code: 6z2v
was solved by
M.Schroeder,
A.Chaikuad,
F.Bracher,
S.Knapp,
Structural Genomicsconsortium (Sgc),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 39.37 / 2.60 |
| Space group | I 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 84.376, 45.393, 106.334, 90.00, 111.16, 90.00 |
| R / Rfree (%) | 20.2 / 26.5 |
Other elements in 6z2v:
The structure of CLK3 A319V Mutant Bound with Beta-Carboline Kh-CARB13 (Cpd 3) also contains other interesting chemical elements:
| Potassium | (K) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the CLK3 A319V Mutant Bound with Beta-Carboline Kh-CARB13 (Cpd 3)
(pdb code 6z2v). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the CLK3 A319V Mutant Bound with Beta-Carboline Kh-CARB13 (Cpd 3), PDB code: 6z2v:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the CLK3 A319V Mutant Bound with Beta-Carboline Kh-CARB13 (Cpd 3), PDB code: 6z2v:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 6z2v
Go back to
Chlorine binding site 1 out
of 2 in the CLK3 A319V Mutant Bound with Beta-Carboline Kh-CARB13 (Cpd 3)
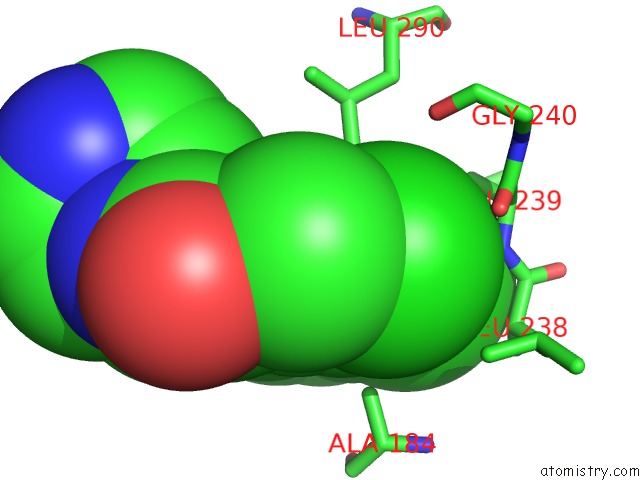
Mono view
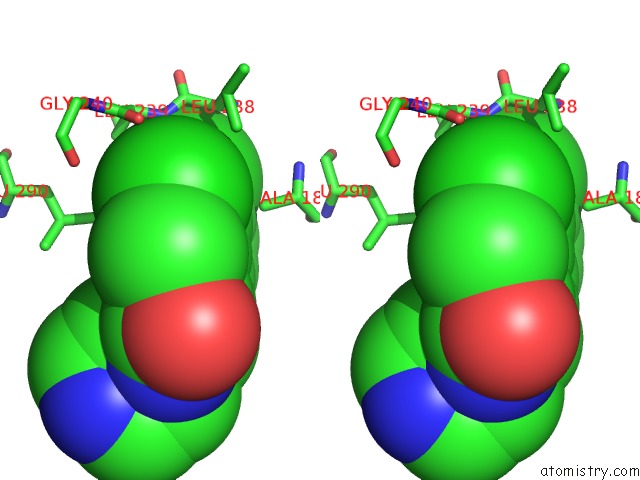
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of CLK3 A319V Mutant Bound with Beta-Carboline Kh-CARB13 (Cpd 3) within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 6z2v
Go back to
Chlorine binding site 2 out
of 2 in the CLK3 A319V Mutant Bound with Beta-Carboline Kh-CARB13 (Cpd 3)
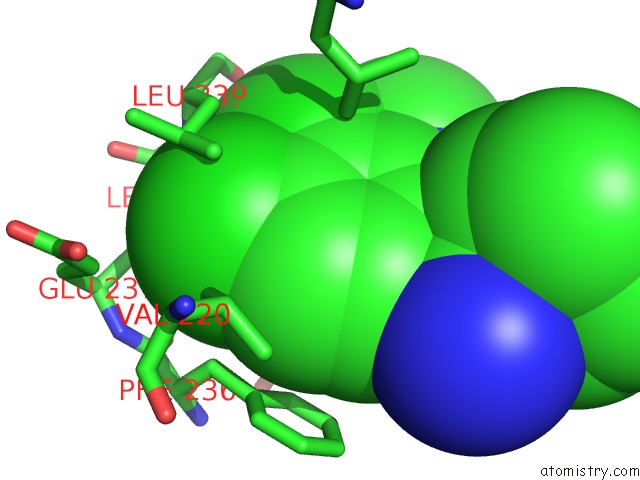
Mono view
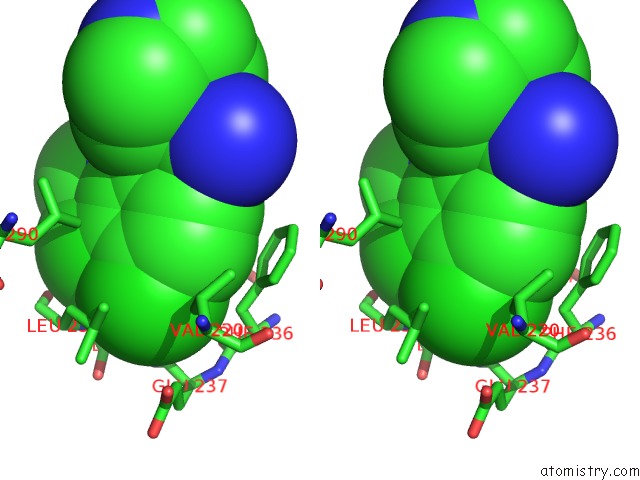
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of CLK3 A319V Mutant Bound with Beta-Carboline Kh-CARB13 (Cpd 3) within 5.0Å range:
|
Reference:
M.Schroder,
A.N.Bullock,
O.Fedorov,
F.Bracher,
A.Chaikuad,
S.Knapp.
Dfg-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing A Basis For Rational Design of Kinase Inhibitor Selectivity. J.Med.Chem. V. 63 10224 2020.
ISSN: ISSN 0022-2623
PubMed: 32787076
DOI: 10.1021/ACS.JMEDCHEM.0C00898
Page generated: Sat Jul 12 22:14:56 2025
ISSN: ISSN 0022-2623
PubMed: 32787076
DOI: 10.1021/ACS.JMEDCHEM.0C00898
Last articles
F in 4IIZF in 4IDQ
F in 4IGH
F in 4IGA
F in 4IBJ
F in 4IFY
F in 4IFV
F in 4IDO
F in 4ICC
F in 4IBI