Chlorine »
PDB 7aq0-7aw2 »
7at9 »
Chlorine in PDB 7at9: Structure of Protein Kinase CK2 Catalytic Subunit (CSNK2A2 Gene Product) in Complex with the Atp-Competitive Inhibitor MB002 and the Alphad-Pocket Ligand 3,4-Dichlorophenethylamine
Enzymatic activity of Structure of Protein Kinase CK2 Catalytic Subunit (CSNK2A2 Gene Product) in Complex with the Atp-Competitive Inhibitor MB002 and the Alphad-Pocket Ligand 3,4-Dichlorophenethylamine
All present enzymatic activity of Structure of Protein Kinase CK2 Catalytic Subunit (CSNK2A2 Gene Product) in Complex with the Atp-Competitive Inhibitor MB002 and the Alphad-Pocket Ligand 3,4-Dichlorophenethylamine:
2.7.11.1;
2.7.11.1;
Protein crystallography data
The structure of Structure of Protein Kinase CK2 Catalytic Subunit (CSNK2A2 Gene Product) in Complex with the Atp-Competitive Inhibitor MB002 and the Alphad-Pocket Ligand 3,4-Dichlorophenethylamine, PDB code: 7at9
was solved by
D.Lindenblatt,
V.Applegate,
A.Nickelsen,
M.Klussmann,
I.Neundorf,
C.Goetz,
J.Jose,
K.Niefind,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 43.74 / 1.05 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 46.482, 47.661, 50.733, 66.61, 89.44, 88.66 |
| R / Rfree (%) | 12.9 / 15 |
Other elements in 7at9:
The structure of Structure of Protein Kinase CK2 Catalytic Subunit (CSNK2A2 Gene Product) in Complex with the Atp-Competitive Inhibitor MB002 and the Alphad-Pocket Ligand 3,4-Dichlorophenethylamine also contains other interesting chemical elements:
| Bromine | (Br) | 8 atoms |
| Sodium | (Na) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Structure of Protein Kinase CK2 Catalytic Subunit (CSNK2A2 Gene Product) in Complex with the Atp-Competitive Inhibitor MB002 and the Alphad-Pocket Ligand 3,4-Dichlorophenethylamine
(pdb code 7at9). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 6 binding sites of Chlorine where determined in the Structure of Protein Kinase CK2 Catalytic Subunit (CSNK2A2 Gene Product) in Complex with the Atp-Competitive Inhibitor MB002 and the Alphad-Pocket Ligand 3,4-Dichlorophenethylamine, PDB code: 7at9:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Chlorine where determined in the Structure of Protein Kinase CK2 Catalytic Subunit (CSNK2A2 Gene Product) in Complex with the Atp-Competitive Inhibitor MB002 and the Alphad-Pocket Ligand 3,4-Dichlorophenethylamine, PDB code: 7at9:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
Chlorine binding site 1 out of 6 in 7at9
Go back to
Chlorine binding site 1 out
of 6 in the Structure of Protein Kinase CK2 Catalytic Subunit (CSNK2A2 Gene Product) in Complex with the Atp-Competitive Inhibitor MB002 and the Alphad-Pocket Ligand 3,4-Dichlorophenethylamine
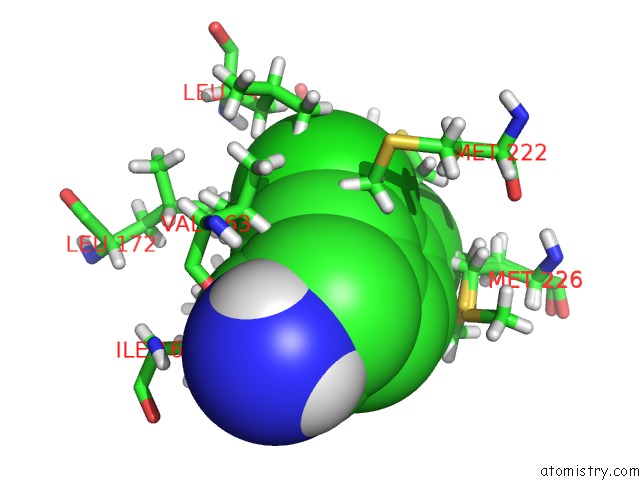
Mono view
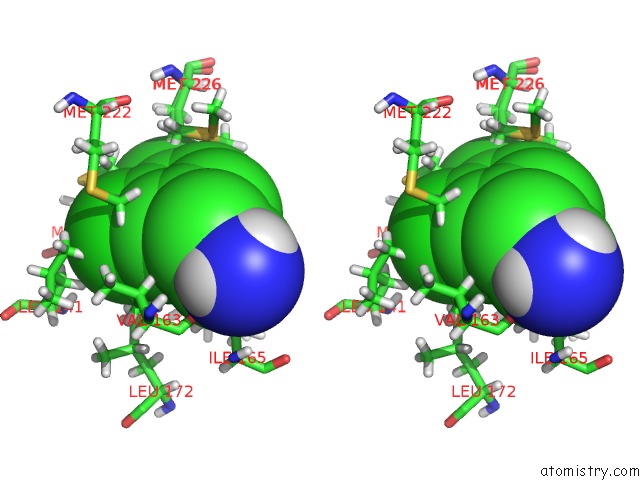
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Structure of Protein Kinase CK2 Catalytic Subunit (CSNK2A2 Gene Product) in Complex with the Atp-Competitive Inhibitor MB002 and the Alphad-Pocket Ligand 3,4-Dichlorophenethylamine within 5.0Å range:
|
Chlorine binding site 2 out of 6 in 7at9
Go back to
Chlorine binding site 2 out
of 6 in the Structure of Protein Kinase CK2 Catalytic Subunit (CSNK2A2 Gene Product) in Complex with the Atp-Competitive Inhibitor MB002 and the Alphad-Pocket Ligand 3,4-Dichlorophenethylamine
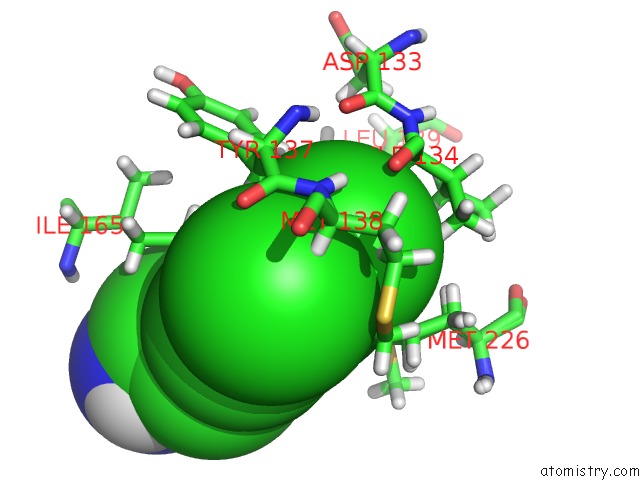
Mono view
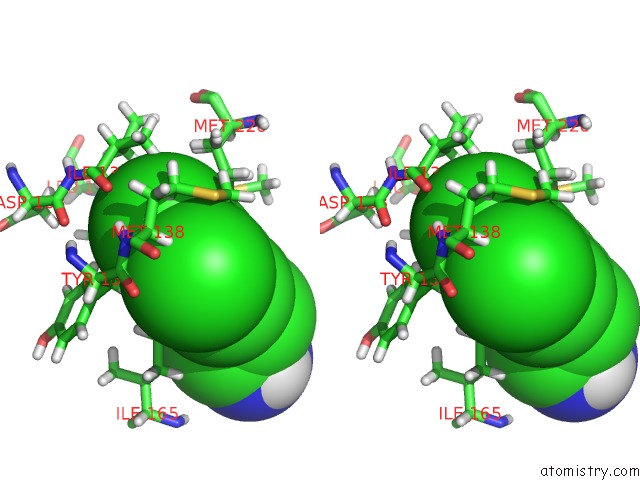
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Structure of Protein Kinase CK2 Catalytic Subunit (CSNK2A2 Gene Product) in Complex with the Atp-Competitive Inhibitor MB002 and the Alphad-Pocket Ligand 3,4-Dichlorophenethylamine within 5.0Å range:
|
Chlorine binding site 3 out of 6 in 7at9
Go back to
Chlorine binding site 3 out
of 6 in the Structure of Protein Kinase CK2 Catalytic Subunit (CSNK2A2 Gene Product) in Complex with the Atp-Competitive Inhibitor MB002 and the Alphad-Pocket Ligand 3,4-Dichlorophenethylamine
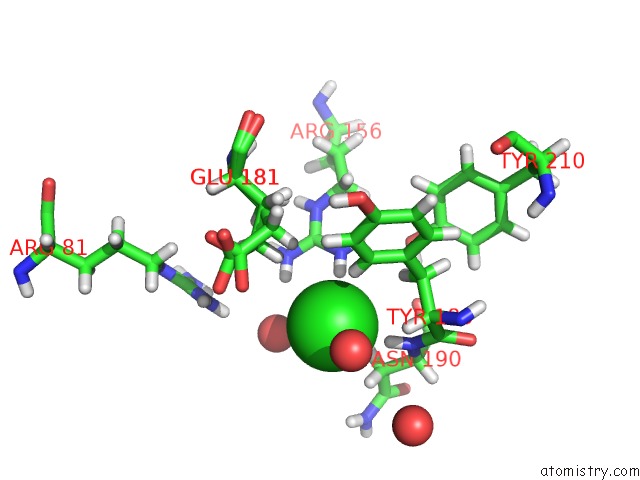
Mono view
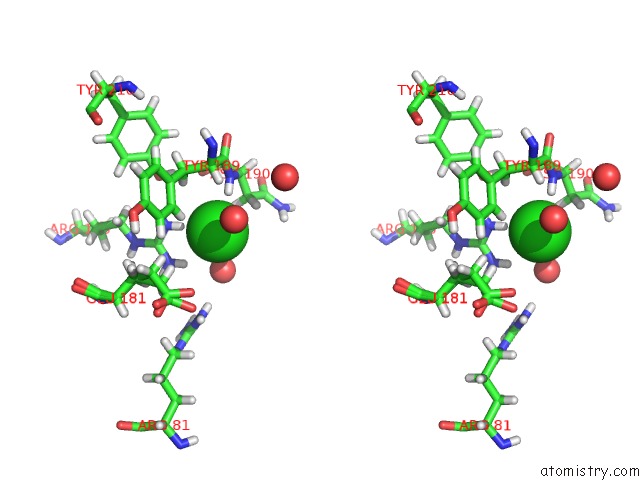
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Structure of Protein Kinase CK2 Catalytic Subunit (CSNK2A2 Gene Product) in Complex with the Atp-Competitive Inhibitor MB002 and the Alphad-Pocket Ligand 3,4-Dichlorophenethylamine within 5.0Å range:
|
Chlorine binding site 4 out of 6 in 7at9
Go back to
Chlorine binding site 4 out
of 6 in the Structure of Protein Kinase CK2 Catalytic Subunit (CSNK2A2 Gene Product) in Complex with the Atp-Competitive Inhibitor MB002 and the Alphad-Pocket Ligand 3,4-Dichlorophenethylamine
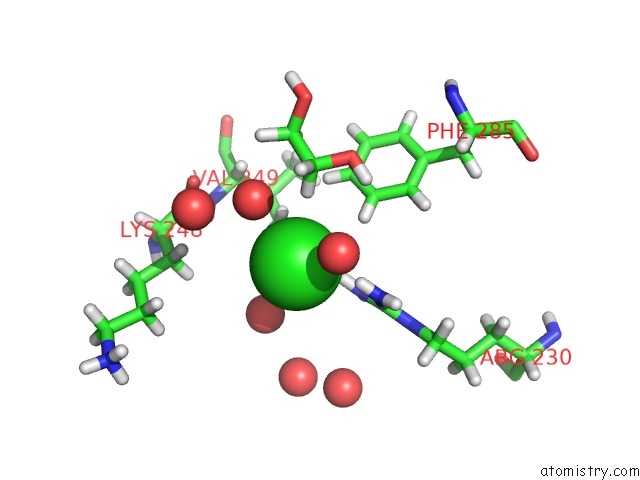
Mono view
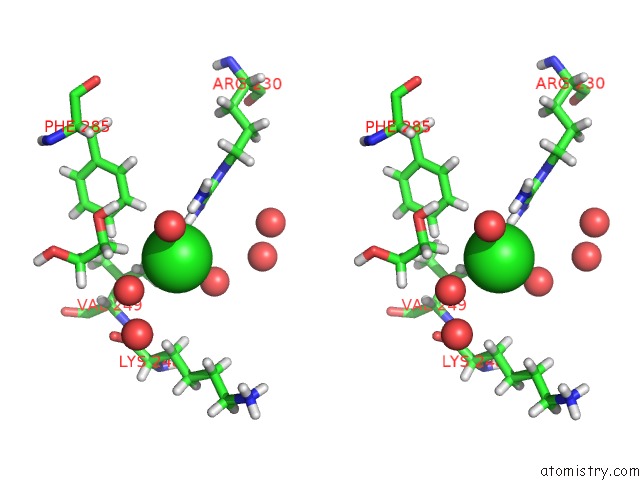
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Structure of Protein Kinase CK2 Catalytic Subunit (CSNK2A2 Gene Product) in Complex with the Atp-Competitive Inhibitor MB002 and the Alphad-Pocket Ligand 3,4-Dichlorophenethylamine within 5.0Å range:
|
Chlorine binding site 5 out of 6 in 7at9
Go back to
Chlorine binding site 5 out
of 6 in the Structure of Protein Kinase CK2 Catalytic Subunit (CSNK2A2 Gene Product) in Complex with the Atp-Competitive Inhibitor MB002 and the Alphad-Pocket Ligand 3,4-Dichlorophenethylamine
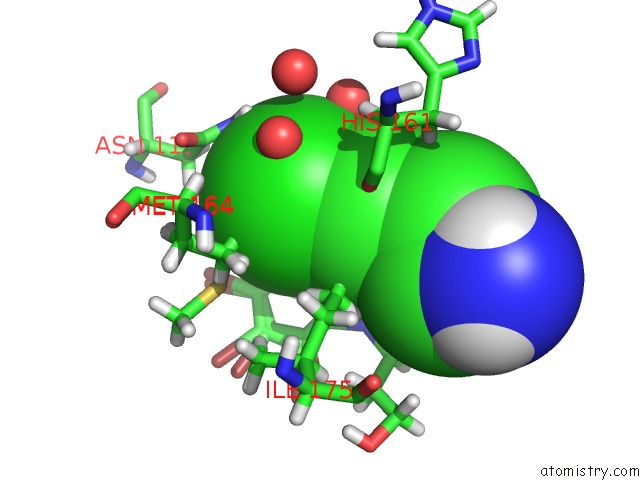
Mono view
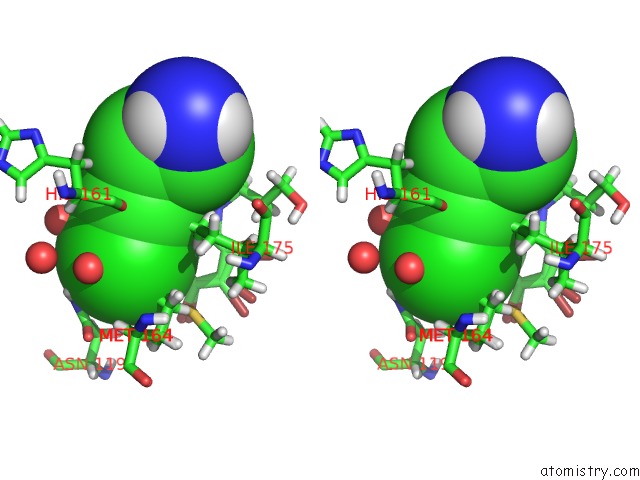
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Structure of Protein Kinase CK2 Catalytic Subunit (CSNK2A2 Gene Product) in Complex with the Atp-Competitive Inhibitor MB002 and the Alphad-Pocket Ligand 3,4-Dichlorophenethylamine within 5.0Å range:
|
Chlorine binding site 6 out of 6 in 7at9
Go back to
Chlorine binding site 6 out
of 6 in the Structure of Protein Kinase CK2 Catalytic Subunit (CSNK2A2 Gene Product) in Complex with the Atp-Competitive Inhibitor MB002 and the Alphad-Pocket Ligand 3,4-Dichlorophenethylamine
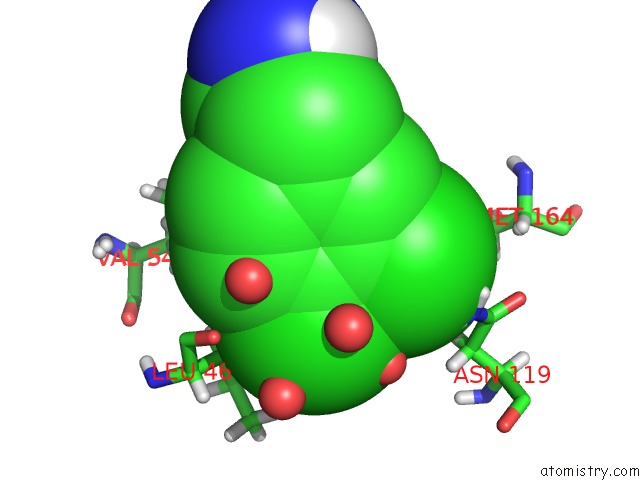
Mono view
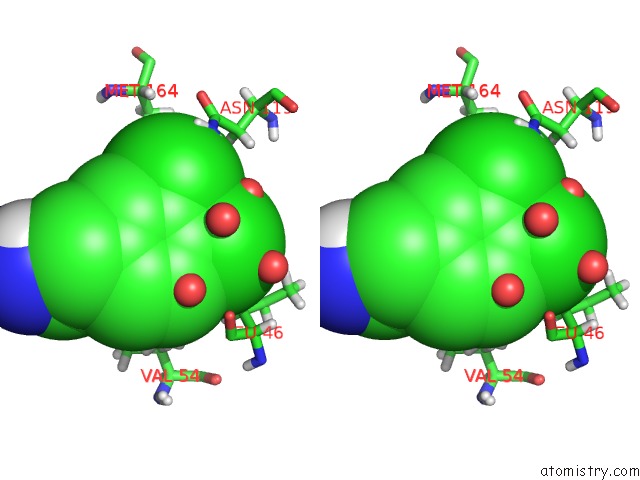
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Structure of Protein Kinase CK2 Catalytic Subunit (CSNK2A2 Gene Product) in Complex with the Atp-Competitive Inhibitor MB002 and the Alphad-Pocket Ligand 3,4-Dichlorophenethylamine within 5.0Å range:
|
Reference:
D.Lindenblatt,
V.Applegate,
A.Nickelsen,
M.Klussmann,
I.Neundorf,
C.Gotz,
J.Jose,
K.Niefind.
Molecular Plasticity of Crystalline CK2 Alpha ' Leads to KN2, A Bivalent Inhibitor of Protein Kinase CK2 with Extraordinary Selectivity J.Med.Chem. 2021.
ISSN: ISSN 0022-2623
DOI: 10.1021/ACS.JMEDCHEM.1C00063
Page generated: Sat Jul 12 22:52:59 2025
ISSN: ISSN 0022-2623
DOI: 10.1021/ACS.JMEDCHEM.1C00063
Last articles
Cl in 8CKPCl in 8CK6
Cl in 8CJF
Cl in 8CJD
Cl in 8CJO
Cl in 8CIP
Cl in 8CJ9
Cl in 8CJ8
Cl in 8CHW
Cl in 8CIG