Chlorine »
PDB 7c74-7ckz »
7cbj »
Chlorine in PDB 7cbj: Crystal Structure of PDE4D Catalytic Domain in Complex with Compound 36
Enzymatic activity of Crystal Structure of PDE4D Catalytic Domain in Complex with Compound 36
All present enzymatic activity of Crystal Structure of PDE4D Catalytic Domain in Complex with Compound 36:
3.1.4.53;
3.1.4.53;
Protein crystallography data
The structure of Crystal Structure of PDE4D Catalytic Domain in Complex with Compound 36, PDB code: 7cbj
was solved by
X.L.Zhang,
Y.C.Xu,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 33.52 / 1.50 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 58.124, 80.463, 164.096, 90, 90, 90 |
| R / Rfree (%) | 18.1 / 20.5 |
Other elements in 7cbj:
The structure of Crystal Structure of PDE4D Catalytic Domain in Complex with Compound 36 also contains other interesting chemical elements:
| Magnesium | (Mg) | 2 atoms |
| Zinc | (Zn) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of PDE4D Catalytic Domain in Complex with Compound 36
(pdb code 7cbj). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Crystal Structure of PDE4D Catalytic Domain in Complex with Compound 36, PDB code: 7cbj:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Crystal Structure of PDE4D Catalytic Domain in Complex with Compound 36, PDB code: 7cbj:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 7cbj
Go back to
Chlorine binding site 1 out
of 2 in the Crystal Structure of PDE4D Catalytic Domain in Complex with Compound 36
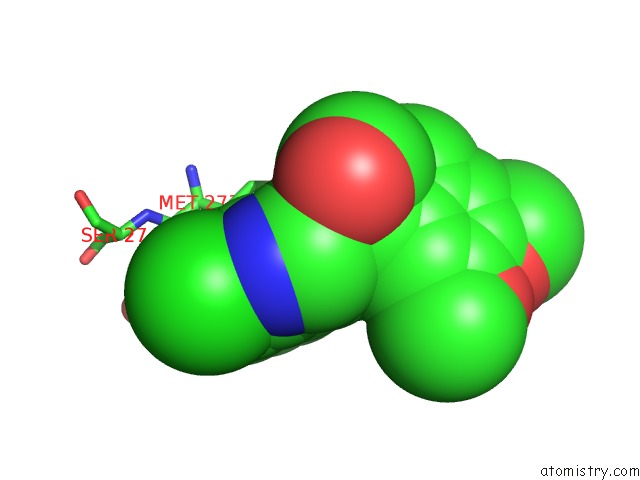
Mono view
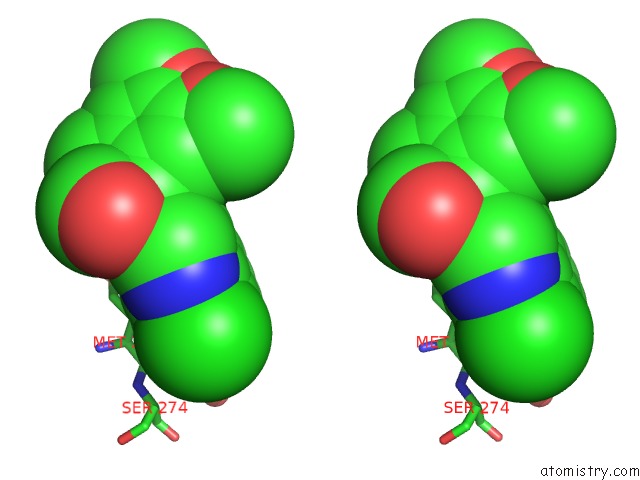
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of PDE4D Catalytic Domain in Complex with Compound 36 within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 7cbj
Go back to
Chlorine binding site 2 out
of 2 in the Crystal Structure of PDE4D Catalytic Domain in Complex with Compound 36
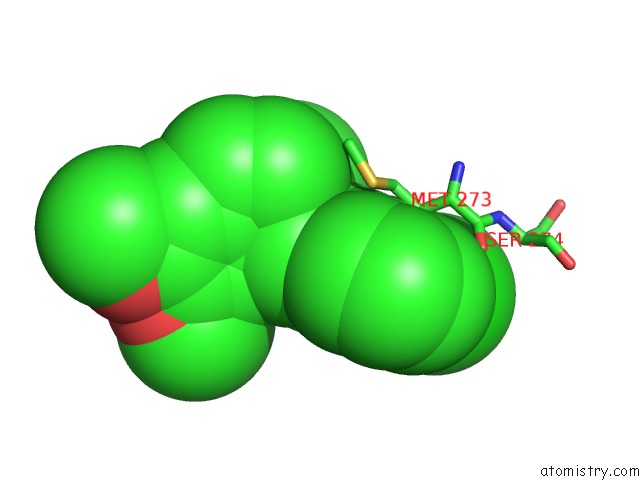
Mono view
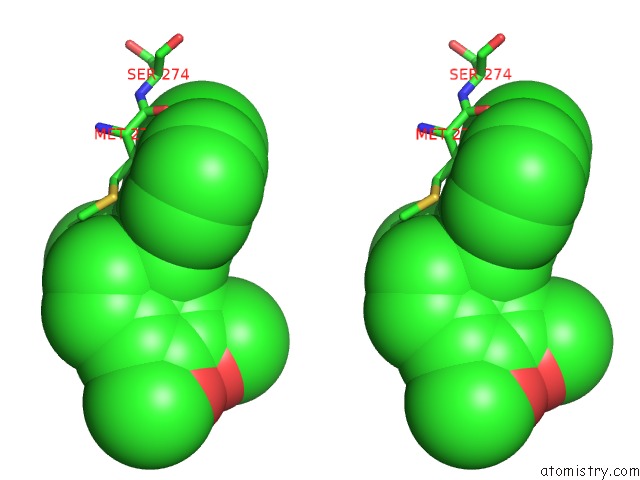
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of PDE4D Catalytic Domain in Complex with Compound 36 within 5.0Å range:
|
Reference:
R.Zhang,
H.Li,
X.Zhang,
J.Li,
H.Su,
Q.Lu,
G.Dong,
H.Dou,
C.Fan,
Z.Gu,
Q.Mu,
W.Tang,
Y.Xu,
H.Liu.
Design, Synthesis, and Biological Evaluation of Tetrahydroisoquinolines Derivatives As Novel, Selective PDE4 Inhibitors For Antipsoriasis Treatment. Eur.J.Med.Chem. V. 211 13004 2021.
ISSN: ISSN 0223-5234
PubMed: 33218684
DOI: 10.1016/J.EJMECH.2020.113004
Page generated: Mon Jul 29 19:39:50 2024
ISSN: ISSN 0223-5234
PubMed: 33218684
DOI: 10.1016/J.EJMECH.2020.113004
Last articles
Cl in 2WOCCl in 2WOD
Cl in 2WM2
Cl in 2WO4
Cl in 2WNY
Cl in 2WNQ
Cl in 2WMZ
Cl in 2WNT
Cl in 2WNS
Cl in 2WLJ