Chlorine »
PDB 7gk8-7glb »
7gkk »
Chlorine in PDB 7gkk: Group Deposition Sars-Cov-2 Main Protease in Complex with Inhibitors From the Covid Moonshot -- Crystal Structure of Sars-Cov-2 Main Protease in Complex with Jin-Pos-6DC588A4-6 (Mpro-P0906)
Enzymatic activity of Group Deposition Sars-Cov-2 Main Protease in Complex with Inhibitors From the Covid Moonshot -- Crystal Structure of Sars-Cov-2 Main Protease in Complex with Jin-Pos-6DC588A4-6 (Mpro-P0906)
All present enzymatic activity of Group Deposition Sars-Cov-2 Main Protease in Complex with Inhibitors From the Covid Moonshot -- Crystal Structure of Sars-Cov-2 Main Protease in Complex with Jin-Pos-6DC588A4-6 (Mpro-P0906):
3.4.22.69;
3.4.22.69;
Protein crystallography data
The structure of Group Deposition Sars-Cov-2 Main Protease in Complex with Inhibitors From the Covid Moonshot -- Crystal Structure of Sars-Cov-2 Main Protease in Complex with Jin-Pos-6DC588A4-6 (Mpro-P0906), PDB code: 7gkk
was solved by
D.Fearon,
A.Aimon,
J.C.Aschenbrenner,
B.H.Balcomb,
F.K.R.Bertram,
J.Brandao-Neto,
A.Dias,
A.Douangamath,
L.Dunnett,
A.S.Godoy,
T.J.Gorrie-Stone,
L.Koekemoer,
T.Krojer,
R.M.Lithgo,
P.Lukacik,
P.G.Marples,
H.Mikolajek,
E.Nelson,
C.D.Owen,
A.J.Powell,
V.L.Rangel,
R.Skyner,
C.M.Strain-Damerell,
W.Thompson,
C.W.E.Tomlinson,
C.Wild,
M.A.Walsh,
F.Von Delft,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 55.65 / 1.80 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 67.27, 99.04, 103.36, 90, 90, 90 |
| R / Rfree (%) | 23.9 / 27.7 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Group Deposition Sars-Cov-2 Main Protease in Complex with Inhibitors From the Covid Moonshot -- Crystal Structure of Sars-Cov-2 Main Protease in Complex with Jin-Pos-6DC588A4-6 (Mpro-P0906)
(pdb code 7gkk). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 3 binding sites of Chlorine where determined in the Group Deposition Sars-Cov-2 Main Protease in Complex with Inhibitors From the Covid Moonshot -- Crystal Structure of Sars-Cov-2 Main Protease in Complex with Jin-Pos-6DC588A4-6 (Mpro-P0906), PDB code: 7gkk:
Jump to Chlorine binding site number: 1; 2; 3;
In total 3 binding sites of Chlorine where determined in the Group Deposition Sars-Cov-2 Main Protease in Complex with Inhibitors From the Covid Moonshot -- Crystal Structure of Sars-Cov-2 Main Protease in Complex with Jin-Pos-6DC588A4-6 (Mpro-P0906), PDB code: 7gkk:
Jump to Chlorine binding site number: 1; 2; 3;
Chlorine binding site 1 out of 3 in 7gkk
Go back to
Chlorine binding site 1 out
of 3 in the Group Deposition Sars-Cov-2 Main Protease in Complex with Inhibitors From the Covid Moonshot -- Crystal Structure of Sars-Cov-2 Main Protease in Complex with Jin-Pos-6DC588A4-6 (Mpro-P0906)
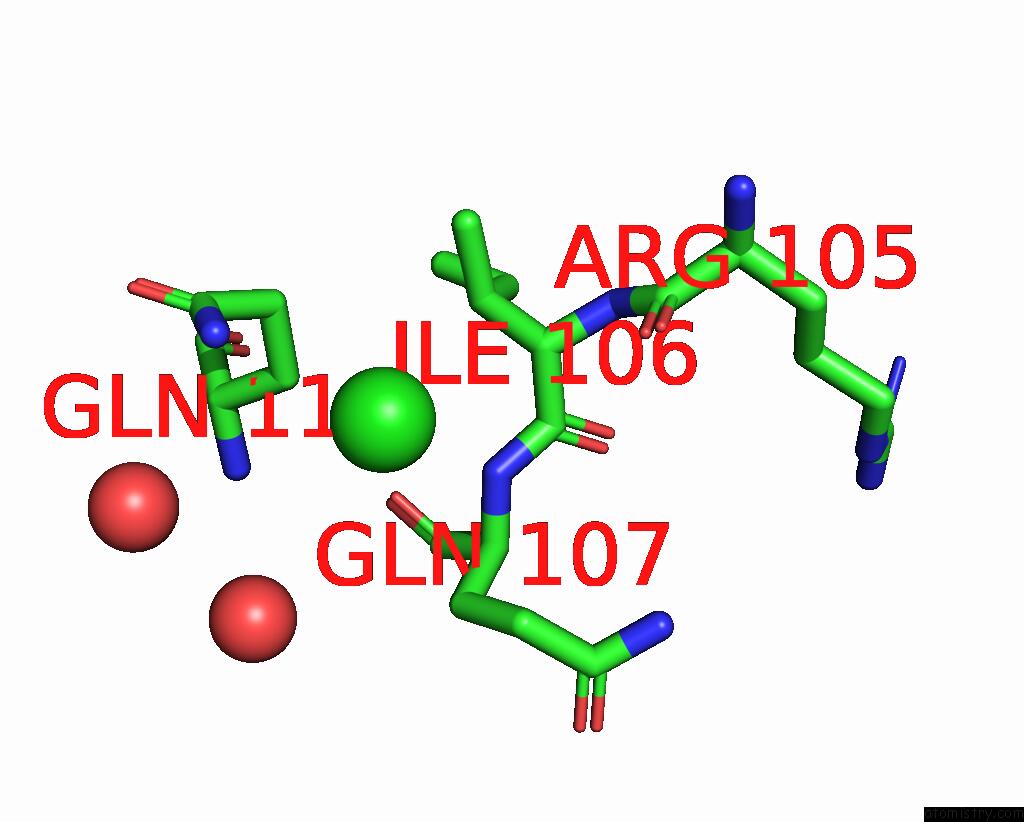
Mono view
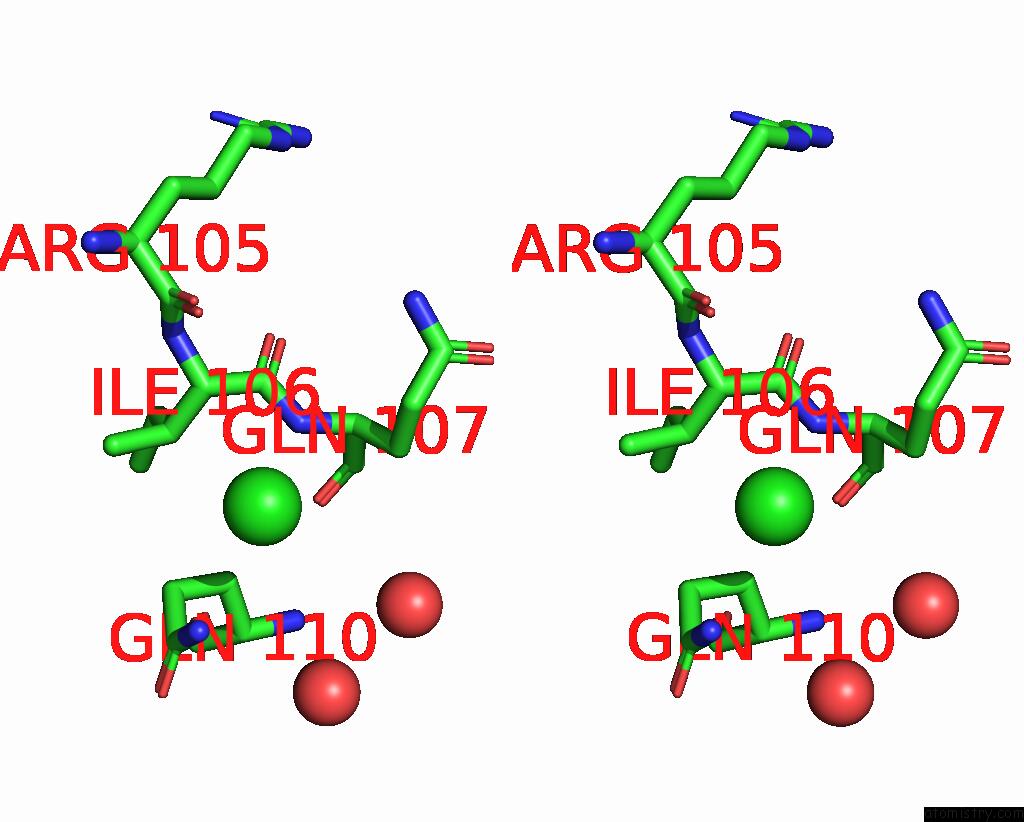
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Group Deposition Sars-Cov-2 Main Protease in Complex with Inhibitors From the Covid Moonshot -- Crystal Structure of Sars-Cov-2 Main Protease in Complex with Jin-Pos-6DC588A4-6 (Mpro-P0906) within 5.0Å range:
|
Chlorine binding site 2 out of 3 in 7gkk
Go back to
Chlorine binding site 2 out
of 3 in the Group Deposition Sars-Cov-2 Main Protease in Complex with Inhibitors From the Covid Moonshot -- Crystal Structure of Sars-Cov-2 Main Protease in Complex with Jin-Pos-6DC588A4-6 (Mpro-P0906)
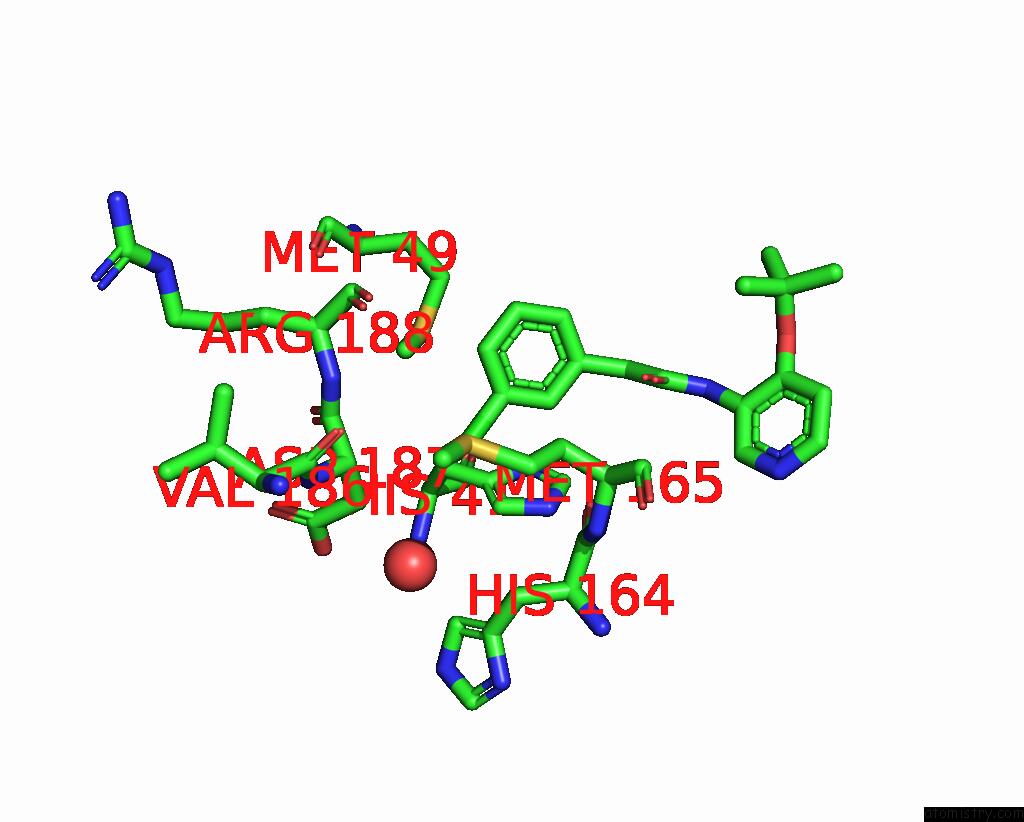
Mono view
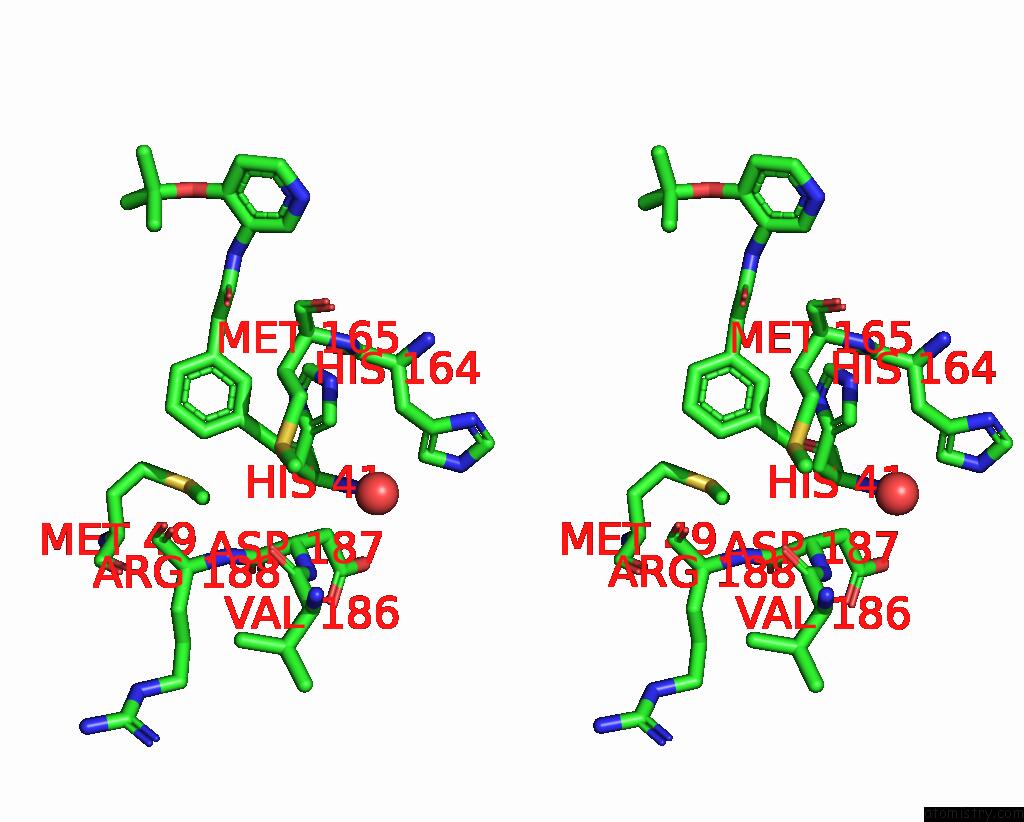
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Group Deposition Sars-Cov-2 Main Protease in Complex with Inhibitors From the Covid Moonshot -- Crystal Structure of Sars-Cov-2 Main Protease in Complex with Jin-Pos-6DC588A4-6 (Mpro-P0906) within 5.0Å range:
|
Chlorine binding site 3 out of 3 in 7gkk
Go back to
Chlorine binding site 3 out
of 3 in the Group Deposition Sars-Cov-2 Main Protease in Complex with Inhibitors From the Covid Moonshot -- Crystal Structure of Sars-Cov-2 Main Protease in Complex with Jin-Pos-6DC588A4-6 (Mpro-P0906)
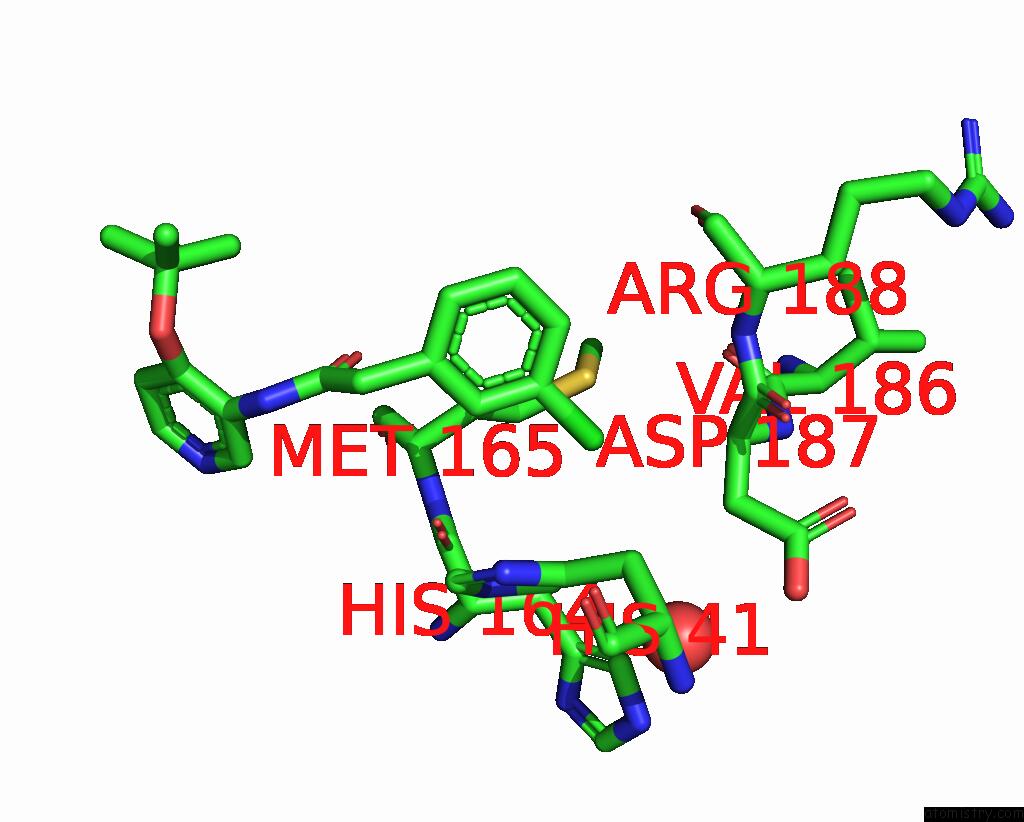
Mono view
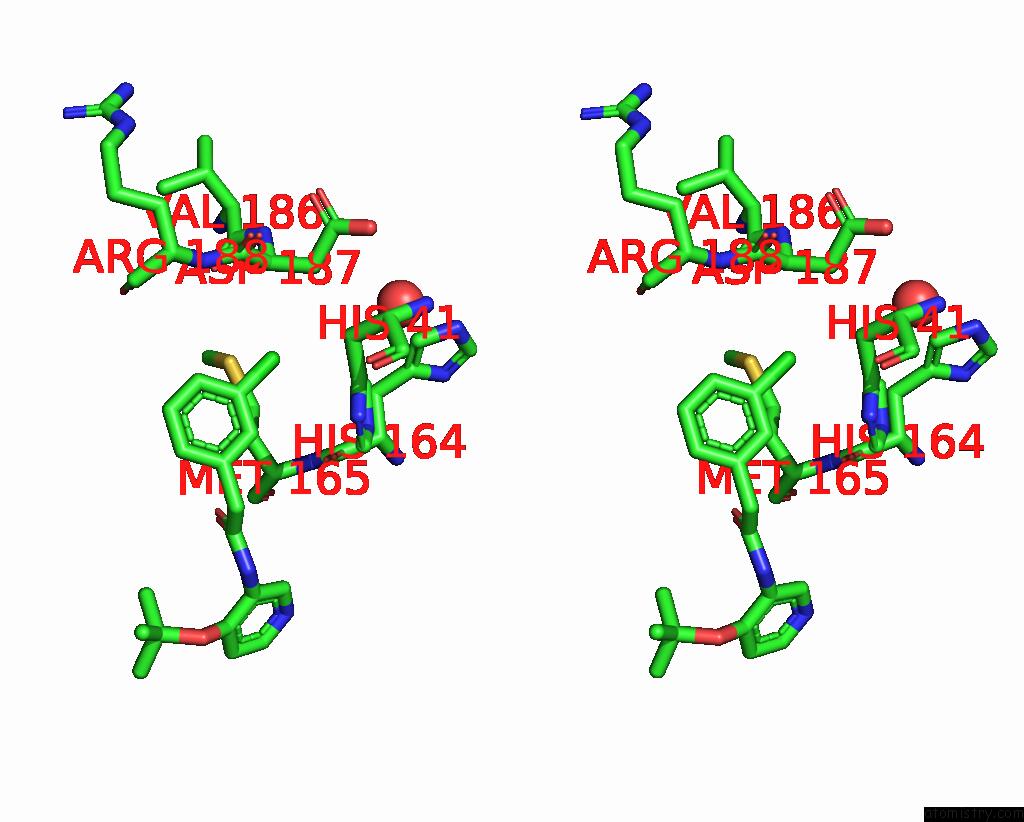
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Group Deposition Sars-Cov-2 Main Protease in Complex with Inhibitors From the Covid Moonshot -- Crystal Structure of Sars-Cov-2 Main Protease in Complex with Jin-Pos-6DC588A4-6 (Mpro-P0906) within 5.0Å range:
|
Reference:
M.L.Boby,
D.Fearon,
M.Ferla,
M.Filep,
L.Koekemoer,
M.C.Robinson,
The Covid Moonshot Consortium,
J.D.Chodera,
A.A.Lee,
N.London,
A.Von Delft,
F.Von Delft.
Open Science Discovery of Potent Noncovalent Sars-Cov-2 Main Protease Inhibitors Science 2023.
ISSN: ESSN 1095-9203
DOI: 10.1126/SCIENCE.ABO7201
Page generated: Sun Jul 13 01:44:30 2025
ISSN: ESSN 1095-9203
DOI: 10.1126/SCIENCE.ABO7201
Last articles
F in 4JVSF in 4JVR
F in 4JU1
F in 4JUO
F in 4JVE
F in 4JU4
F in 4JTZ
F in 4JTY
F in 4JTW
F in 4JSX