Chlorine »
PDB 7obe-7okc »
7ocp »
Chlorine in PDB 7ocp: Nadph Bound to the Dehydrogenase Domain of the Bifunctional Mannitol- 1-Phosphate Dehydrogenase/Phosphatase Mtld From Acinetobacter Baumannii
Protein crystallography data
The structure of Nadph Bound to the Dehydrogenase Domain of the Bifunctional Mannitol- 1-Phosphate Dehydrogenase/Phosphatase Mtld From Acinetobacter Baumannii, PDB code: 7ocp
was solved by
H.K.Tam,
V.Mueller,
K.M.Pos,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.59 / 2.75 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 99.661, 157.692, 219.289, 90, 90, 90 |
| R / Rfree (%) | 22 / 25.5 |
Other elements in 7ocp:
The structure of Nadph Bound to the Dehydrogenase Domain of the Bifunctional Mannitol- 1-Phosphate Dehydrogenase/Phosphatase Mtld From Acinetobacter Baumannii also contains other interesting chemical elements:
| Magnesium | (Mg) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Nadph Bound to the Dehydrogenase Domain of the Bifunctional Mannitol- 1-Phosphate Dehydrogenase/Phosphatase Mtld From Acinetobacter Baumannii
(pdb code 7ocp). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Nadph Bound to the Dehydrogenase Domain of the Bifunctional Mannitol- 1-Phosphate Dehydrogenase/Phosphatase Mtld From Acinetobacter Baumannii, PDB code: 7ocp:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Nadph Bound to the Dehydrogenase Domain of the Bifunctional Mannitol- 1-Phosphate Dehydrogenase/Phosphatase Mtld From Acinetobacter Baumannii, PDB code: 7ocp:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 7ocp
Go back to
Chlorine binding site 1 out
of 2 in the Nadph Bound to the Dehydrogenase Domain of the Bifunctional Mannitol- 1-Phosphate Dehydrogenase/Phosphatase Mtld From Acinetobacter Baumannii
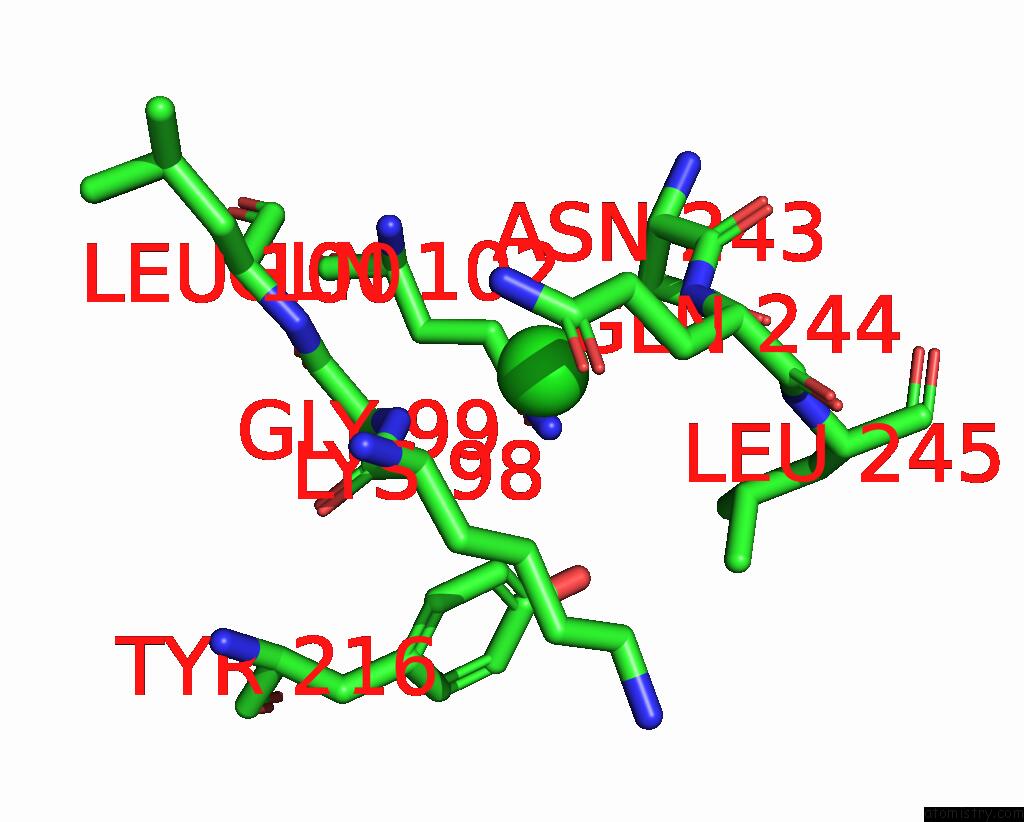
Mono view
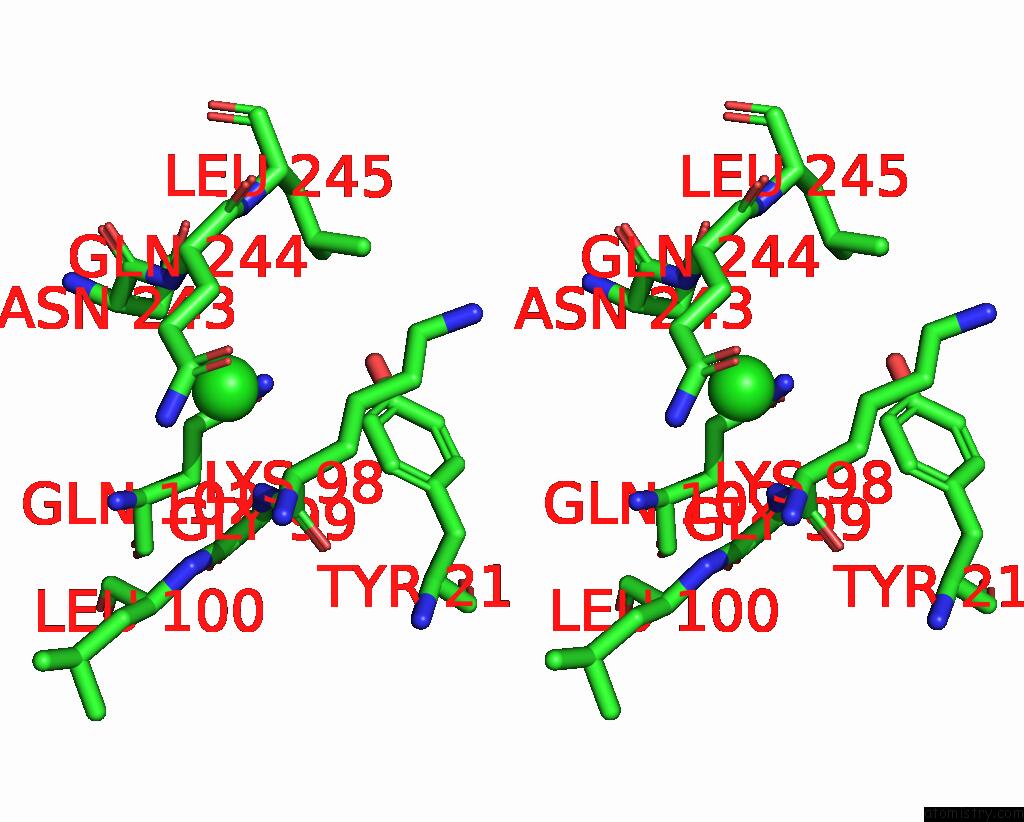
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Nadph Bound to the Dehydrogenase Domain of the Bifunctional Mannitol- 1-Phosphate Dehydrogenase/Phosphatase Mtld From Acinetobacter Baumannii within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 7ocp
Go back to
Chlorine binding site 2 out
of 2 in the Nadph Bound to the Dehydrogenase Domain of the Bifunctional Mannitol- 1-Phosphate Dehydrogenase/Phosphatase Mtld From Acinetobacter Baumannii
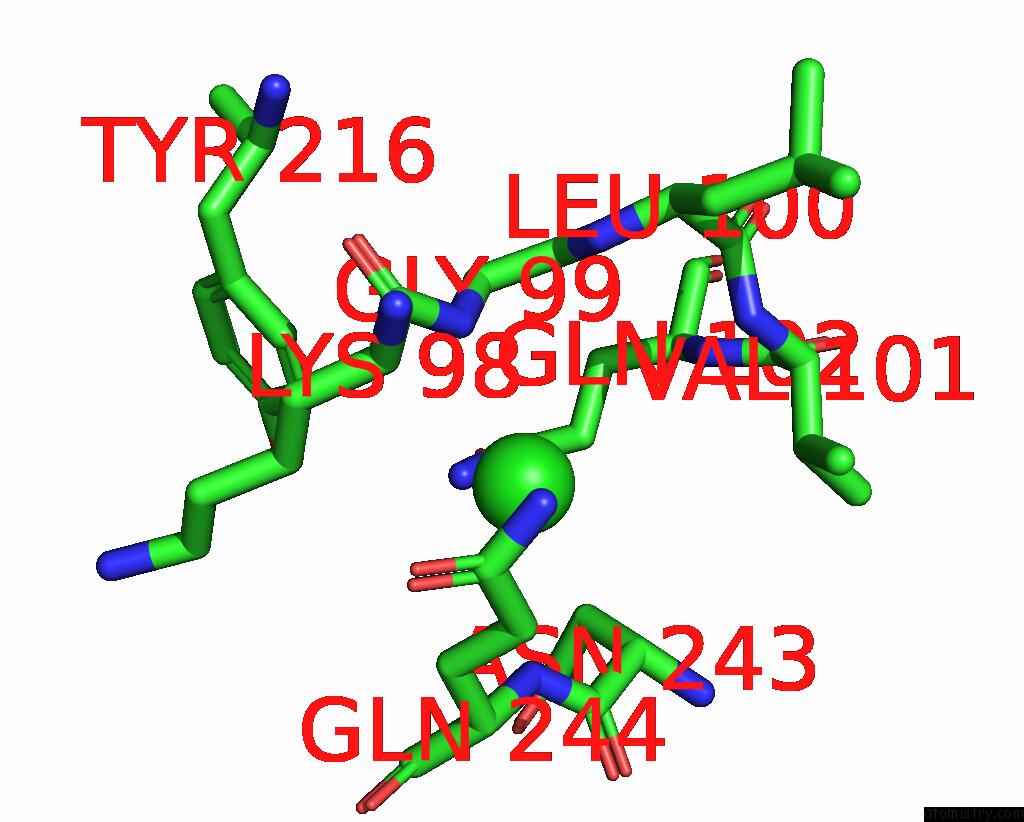
Mono view
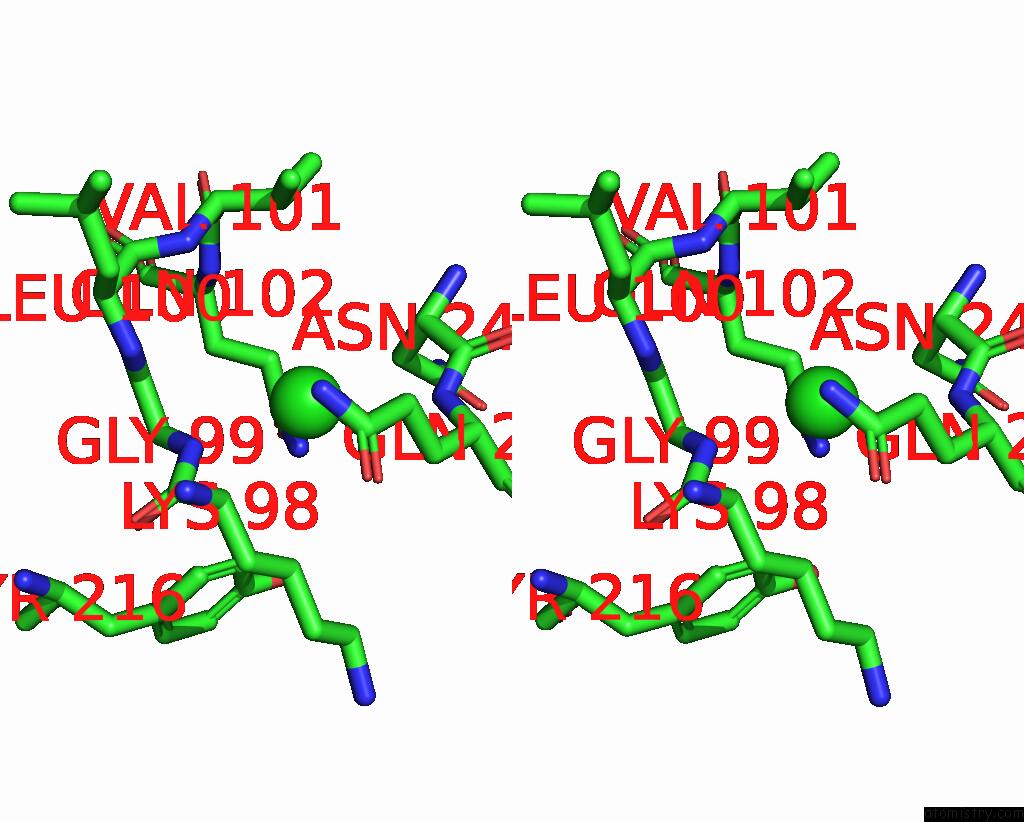
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Nadph Bound to the Dehydrogenase Domain of the Bifunctional Mannitol- 1-Phosphate Dehydrogenase/Phosphatase Mtld From Acinetobacter Baumannii within 5.0Å range:
|
Reference:
H.K.Tam,
P.Konig,
S.Himpich,
N.D.Ngu,
R.Abele,
V.Muller,
K.M.Pos.
Unidirectional Mannitol Synthesis of Acinetobacter Baumannii Mtld Is Facilitated By the Helix-Loop-Helix-Mediated Dimer Formation. Proc.Natl.Acad.Sci.Usa V. 119 94119 2022.
ISSN: ESSN 1091-6490
PubMed: 35363566
DOI: 10.1073/PNAS.2107994119
Page generated: Sun Jul 13 04:52:52 2025
ISSN: ESSN 1091-6490
PubMed: 35363566
DOI: 10.1073/PNAS.2107994119
Last articles
Cl in 8OJWCl in 8OJP
Cl in 8OJX
Cl in 8OJI
Cl in 8OJV
Cl in 8OI4
Cl in 8OJF
Cl in 8OIG
Cl in 8OJ2
Cl in 8OJ1