Chlorine »
PDB 7okd-7ooz »
7ood »
Chlorine in PDB 7ood: Mycoplasma Pneumoniae 50S Subunit of Ribosomes in Chloramphenicol- Treated Cells
Other elements in 7ood:
The structure of Mycoplasma Pneumoniae 50S Subunit of Ribosomes in Chloramphenicol- Treated Cells also contains other interesting chemical elements:
| Zinc | (Zn) | 3 atoms |
| Magnesium | (Mg) | 25 atoms |
| Potassium | (K) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Mycoplasma Pneumoniae 50S Subunit of Ribosomes in Chloramphenicol- Treated Cells
(pdb code 7ood). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Mycoplasma Pneumoniae 50S Subunit of Ribosomes in Chloramphenicol- Treated Cells, PDB code: 7ood:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Mycoplasma Pneumoniae 50S Subunit of Ribosomes in Chloramphenicol- Treated Cells, PDB code: 7ood:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 7ood
Go back to
Chlorine binding site 1 out
of 2 in the Mycoplasma Pneumoniae 50S Subunit of Ribosomes in Chloramphenicol- Treated Cells
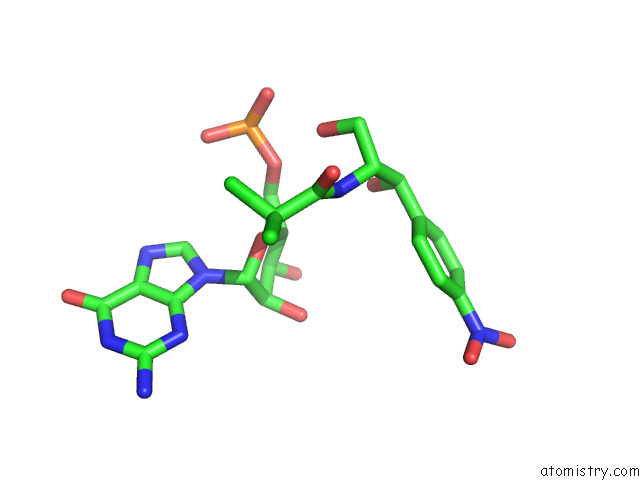
Mono view
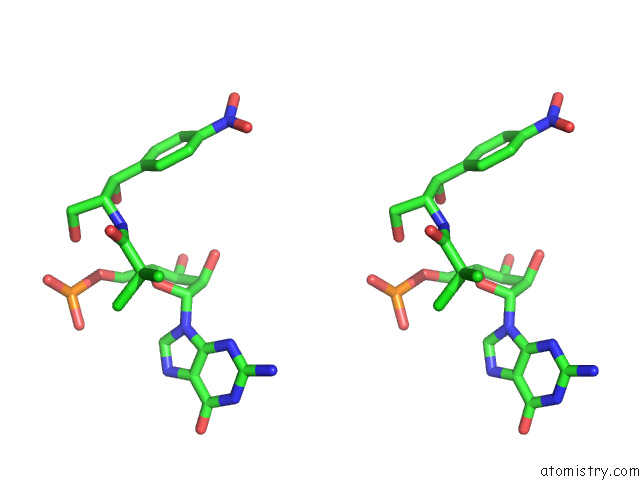
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Mycoplasma Pneumoniae 50S Subunit of Ribosomes in Chloramphenicol- Treated Cells within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 7ood
Go back to
Chlorine binding site 2 out
of 2 in the Mycoplasma Pneumoniae 50S Subunit of Ribosomes in Chloramphenicol- Treated Cells
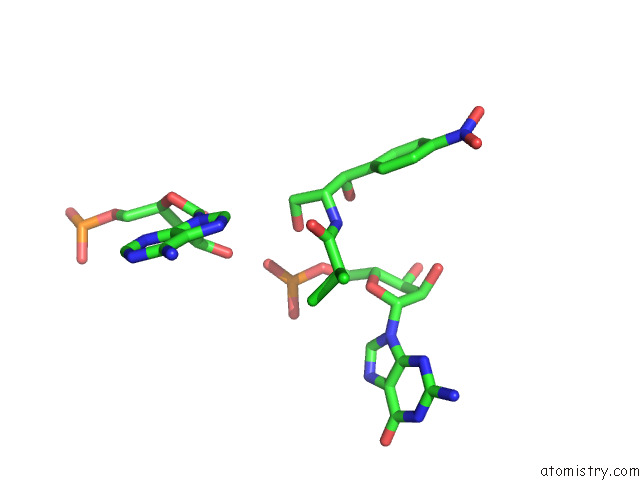
Mono view
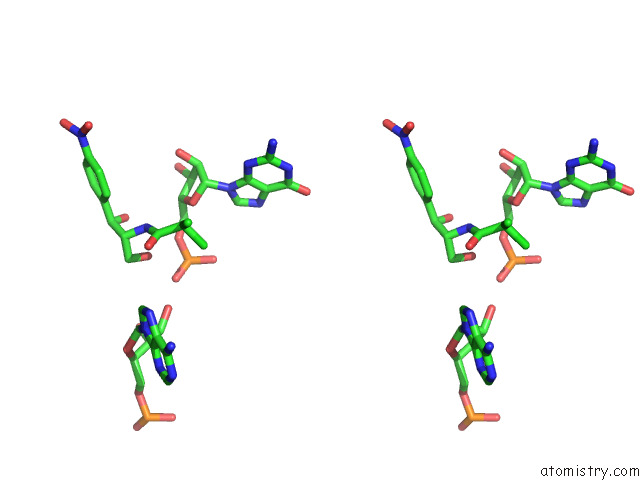
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Mycoplasma Pneumoniae 50S Subunit of Ribosomes in Chloramphenicol- Treated Cells within 5.0Å range:
|
Reference:
L.Xue,
S.Lenz,
M.Zimmermann-Kogadeeva,
D.Tegunov,
P.Cramer,
P.Bork,
J.Rappsilber,
J.Mahamid.
Visualizing Translation Dynamics at Atomic Detail Inside A Bacterial Cell. Nature V. 610 205 2022.
ISSN: ESSN 1476-4687
PubMed: 36171285
DOI: 10.1038/S41586-022-05255-2
Page generated: Sun Jul 13 05:18:35 2025
ISSN: ESSN 1476-4687
PubMed: 36171285
DOI: 10.1038/S41586-022-05255-2
Last articles
F in 7QV9F in 7QSC
F in 7QTM
F in 7QTV
F in 7QU0
F in 7QRA
F in 7QRB
F in 7QRC
F in 7QR9
F in 7QQ6