Chlorine »
PDB 7q1o-7q7m »
7q3r »
Chlorine in PDB 7q3r: Crystal Structure of Sars-Cov-2 Rbd in Complex with the Neutralizing Nanobodies Vhh-F04 and Vhh-G09
Protein crystallography data
The structure of Crystal Structure of Sars-Cov-2 Rbd in Complex with the Neutralizing Nanobodies Vhh-F04 and Vhh-G09, PDB code: 7q3r
was solved by
I.Fernandez,
R.Pederzoli,
F.A.Rey,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.91 / 1.92 |
| Space group | P 63 |
| Cell size a, b, c (Å), α, β, γ (°) | 180.578, 180.578, 48.025, 90, 90, 120 |
| R / Rfree (%) | 18.2 / 20.9 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Sars-Cov-2 Rbd in Complex with the Neutralizing Nanobodies Vhh-F04 and Vhh-G09
(pdb code 7q3r). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 5 binding sites of Chlorine where determined in the Crystal Structure of Sars-Cov-2 Rbd in Complex with the Neutralizing Nanobodies Vhh-F04 and Vhh-G09, PDB code: 7q3r:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Chlorine where determined in the Crystal Structure of Sars-Cov-2 Rbd in Complex with the Neutralizing Nanobodies Vhh-F04 and Vhh-G09, PDB code: 7q3r:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5;
Chlorine binding site 1 out of 5 in 7q3r
Go back to
Chlorine binding site 1 out
of 5 in the Crystal Structure of Sars-Cov-2 Rbd in Complex with the Neutralizing Nanobodies Vhh-F04 and Vhh-G09
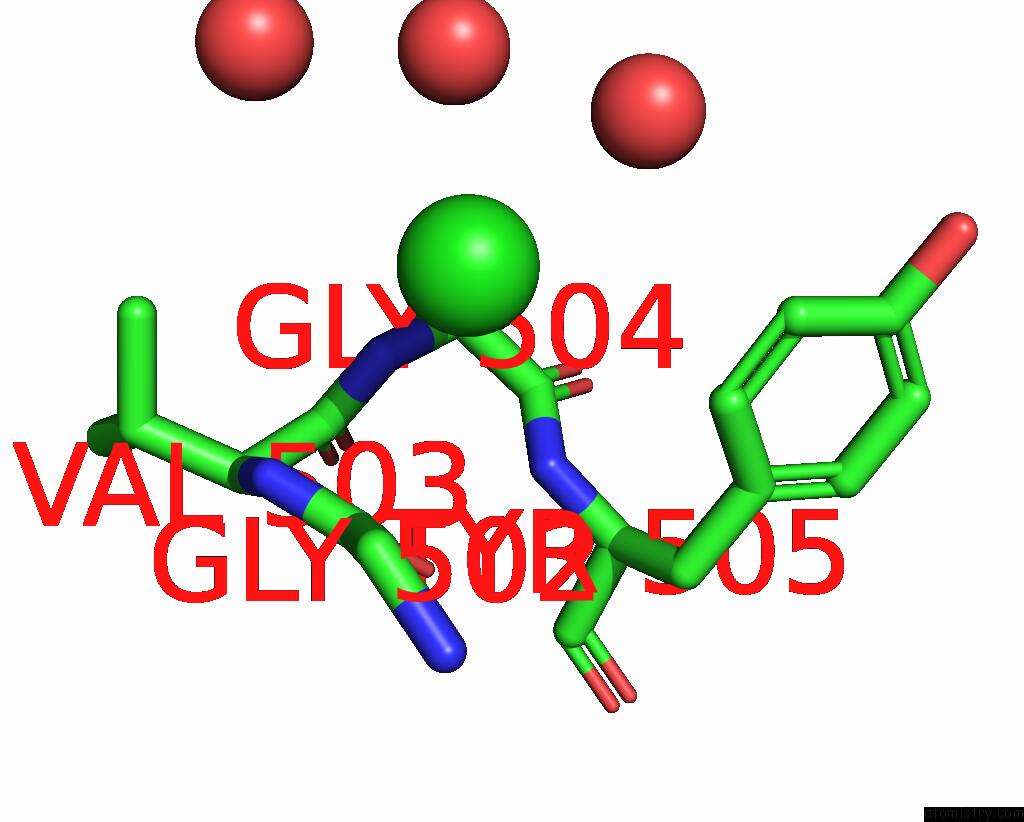
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Sars-Cov-2 Rbd in Complex with the Neutralizing Nanobodies Vhh-F04 and Vhh-G09 within 5.0Å range:
|
Chlorine binding site 2 out of 5 in 7q3r
Go back to
Chlorine binding site 2 out
of 5 in the Crystal Structure of Sars-Cov-2 Rbd in Complex with the Neutralizing Nanobodies Vhh-F04 and Vhh-G09
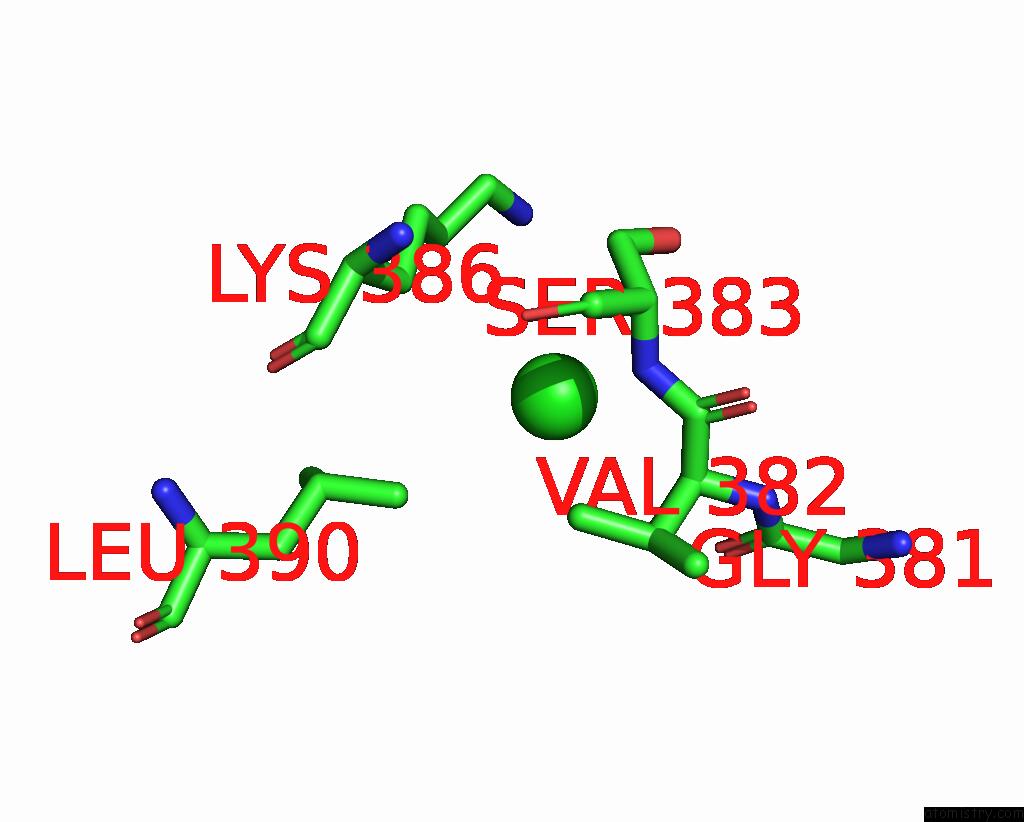
Mono view
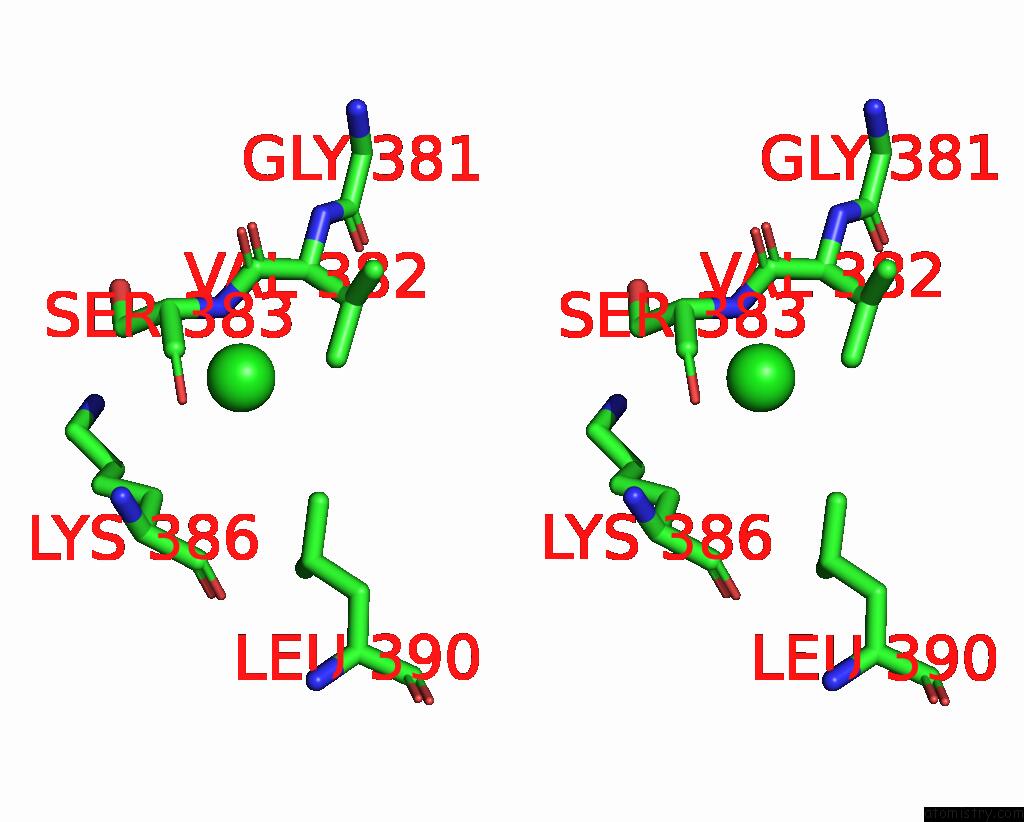
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Sars-Cov-2 Rbd in Complex with the Neutralizing Nanobodies Vhh-F04 and Vhh-G09 within 5.0Å range:
|
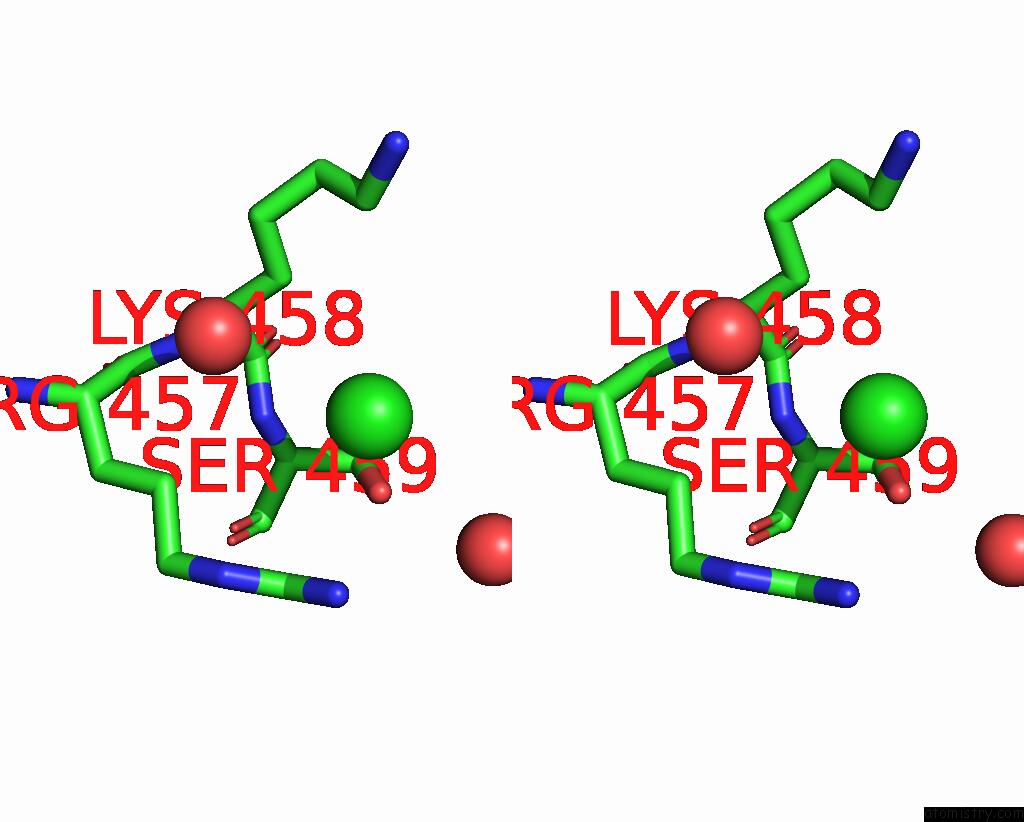
Chlorine binding site 3 out of 5 in 7q3r
Go back to
Chlorine binding site 3 out
of 5 in the Crystal Structure of Sars-Cov-2 Rbd in Complex with the Neutralizing Nanobodies Vhh-F04 and Vhh-G09
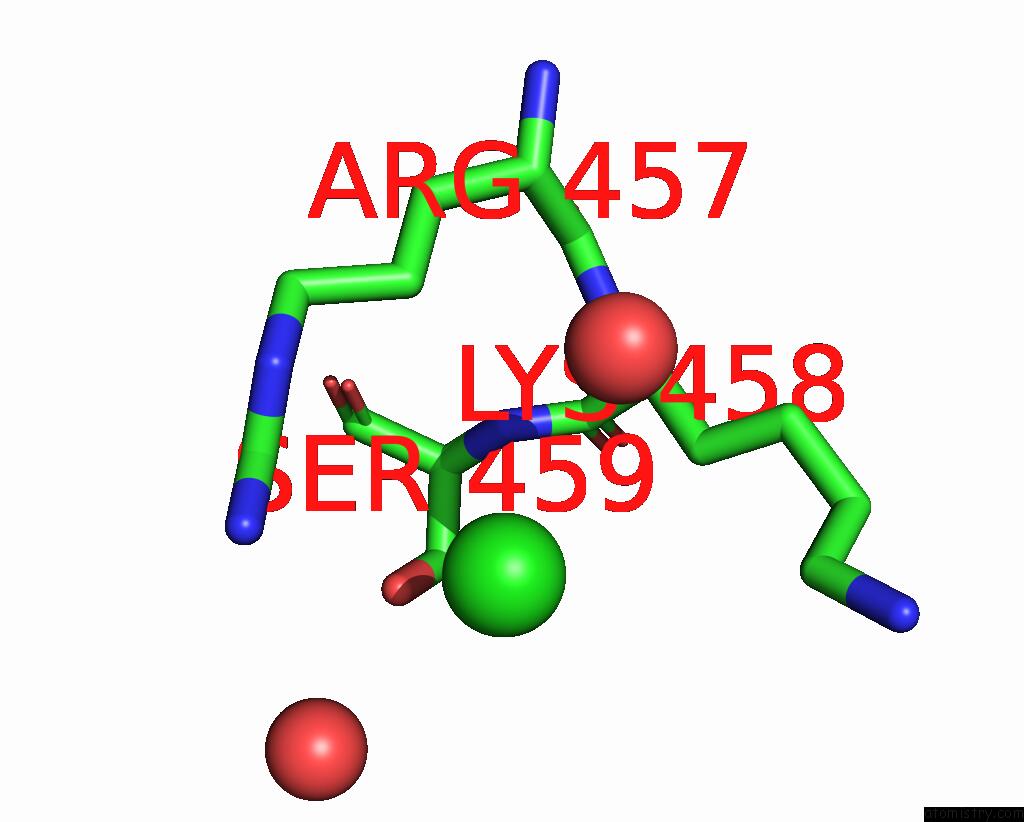
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of Sars-Cov-2 Rbd in Complex with the Neutralizing Nanobodies Vhh-F04 and Vhh-G09 within 5.0Å range:
|
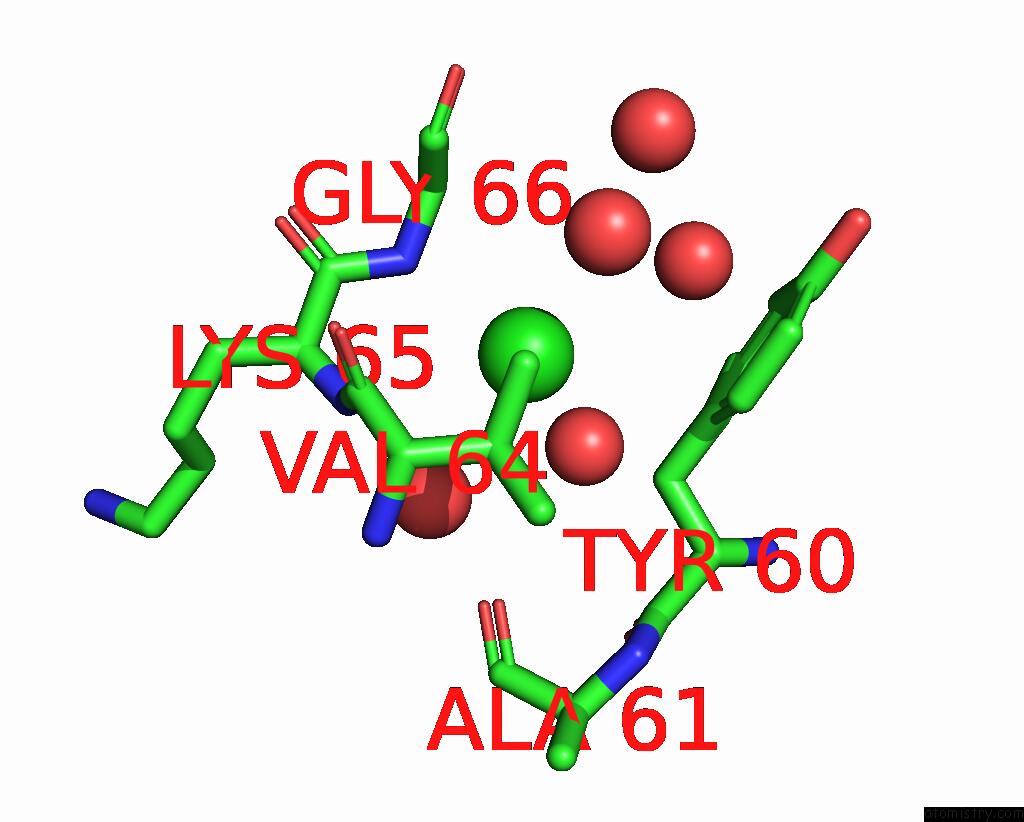
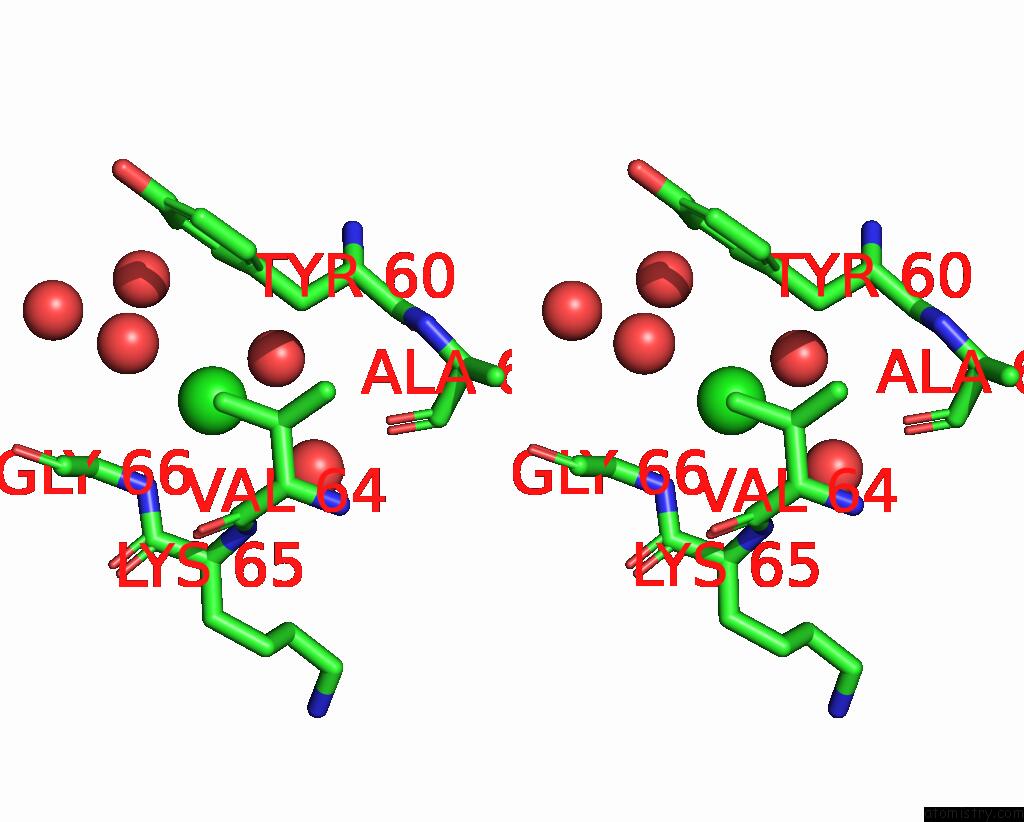
Chlorine binding site 4 out of 5 in 7q3r
Go back to
Chlorine binding site 4 out
of 5 in the Crystal Structure of Sars-Cov-2 Rbd in Complex with the Neutralizing Nanobodies Vhh-F04 and Vhh-G09
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of Sars-Cov-2 Rbd in Complex with the Neutralizing Nanobodies Vhh-F04 and Vhh-G09 within 5.0Å range:
|
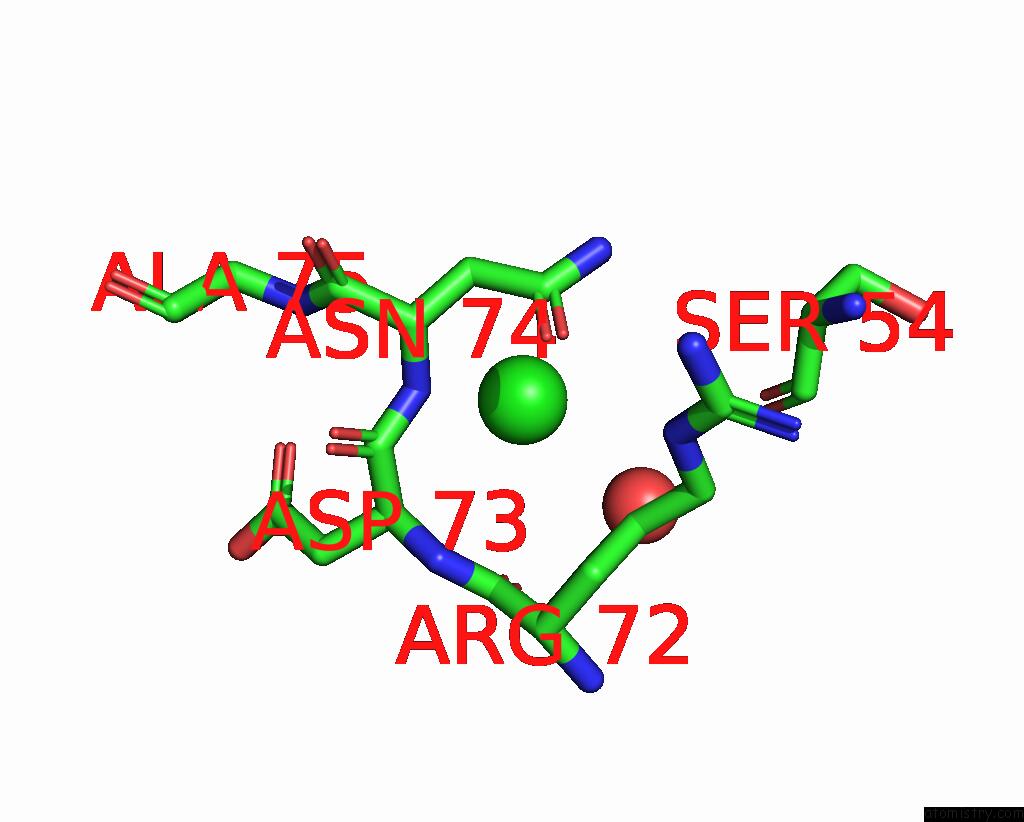
Chlorine binding site 5 out of 5 in 7q3r
Go back to
Chlorine binding site 5 out
of 5 in the Crystal Structure of Sars-Cov-2 Rbd in Complex with the Neutralizing Nanobodies Vhh-F04 and Vhh-G09
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Crystal Structure of Sars-Cov-2 Rbd in Complex with the Neutralizing Nanobodies Vhh-F04 and Vhh-G09 within 5.0Å range:
|
Reference:
I.Fernandez,
F.A.Rey.
Nanobodies Against Sars-Cov-2 Neutralize Variants of Concern and Explore Conformational Differences on the Spike Protein To Be Published.
Page generated: Sun Jul 13 06:09:29 2025
Last articles
Cl in 8SSNCl in 8STN
Cl in 8SR2
Cl in 8SSJ
Cl in 8STB
Cl in 8SQR
Cl in 8SRX
Cl in 8SQT
Cl in 8SQQ
Cl in 8SMQ