Chlorine »
PDB 7sqe-7t2x »
7svt »
Chlorine in PDB 7svt: Mycobacterium Tuberculosis 3-Hydroxyl-Acp Dehydratase Hadab in Complex with 1,3-Diarylpyrazolyl-Acylsulfonamide Inhibitor
Protein crystallography data
The structure of Mycobacterium Tuberculosis 3-Hydroxyl-Acp Dehydratase Hadab in Complex with 1,3-Diarylpyrazolyl-Acylsulfonamide Inhibitor, PDB code: 7svt
was solved by
I.V.Krieger,
J.C.Sacchettini,
Tb Structural Genomics Consortium (Tbsgc),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.38 / 2.40 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 85.298, 107.197, 142.066, 90, 90, 90 |
| R / Rfree (%) | 21.7 / 27.5 |
Other elements in 7svt:
The structure of Mycobacterium Tuberculosis 3-Hydroxyl-Acp Dehydratase Hadab in Complex with 1,3-Diarylpyrazolyl-Acylsulfonamide Inhibitor also contains other interesting chemical elements:
| Bromine | (Br) | 4 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Mycobacterium Tuberculosis 3-Hydroxyl-Acp Dehydratase Hadab in Complex with 1,3-Diarylpyrazolyl-Acylsulfonamide Inhibitor
(pdb code 7svt). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 4 binding sites of Chlorine where determined in the Mycobacterium Tuberculosis 3-Hydroxyl-Acp Dehydratase Hadab in Complex with 1,3-Diarylpyrazolyl-Acylsulfonamide Inhibitor, PDB code: 7svt:
Jump to Chlorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Chlorine where determined in the Mycobacterium Tuberculosis 3-Hydroxyl-Acp Dehydratase Hadab in Complex with 1,3-Diarylpyrazolyl-Acylsulfonamide Inhibitor, PDB code: 7svt:
Jump to Chlorine binding site number: 1; 2; 3; 4;
Chlorine binding site 1 out of 4 in 7svt
Go back to
Chlorine binding site 1 out
of 4 in the Mycobacterium Tuberculosis 3-Hydroxyl-Acp Dehydratase Hadab in Complex with 1,3-Diarylpyrazolyl-Acylsulfonamide Inhibitor
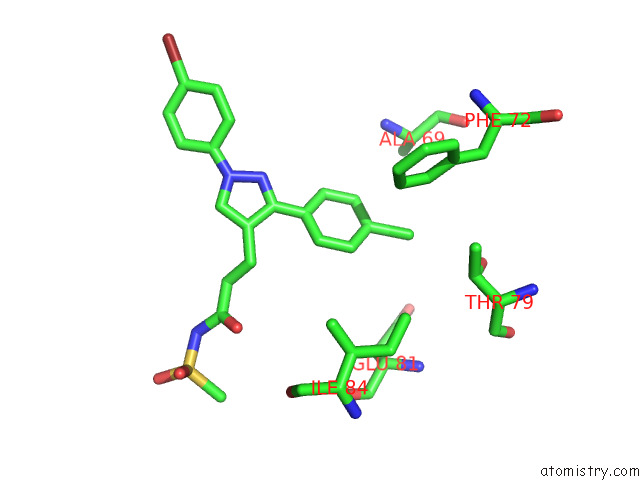
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Mycobacterium Tuberculosis 3-Hydroxyl-Acp Dehydratase Hadab in Complex with 1,3-Diarylpyrazolyl-Acylsulfonamide Inhibitor within 5.0Å range:
|
Chlorine binding site 2 out of 4 in 7svt
Go back to
Chlorine binding site 2 out
of 4 in the Mycobacterium Tuberculosis 3-Hydroxyl-Acp Dehydratase Hadab in Complex with 1,3-Diarylpyrazolyl-Acylsulfonamide Inhibitor

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Mycobacterium Tuberculosis 3-Hydroxyl-Acp Dehydratase Hadab in Complex with 1,3-Diarylpyrazolyl-Acylsulfonamide Inhibitor within 5.0Å range:
|
Chlorine binding site 3 out of 4 in 7svt
Go back to
Chlorine binding site 3 out
of 4 in the Mycobacterium Tuberculosis 3-Hydroxyl-Acp Dehydratase Hadab in Complex with 1,3-Diarylpyrazolyl-Acylsulfonamide Inhibitor

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Mycobacterium Tuberculosis 3-Hydroxyl-Acp Dehydratase Hadab in Complex with 1,3-Diarylpyrazolyl-Acylsulfonamide Inhibitor within 5.0Å range:
|
Chlorine binding site 4 out of 4 in 7svt
Go back to
Chlorine binding site 4 out
of 4 in the Mycobacterium Tuberculosis 3-Hydroxyl-Acp Dehydratase Hadab in Complex with 1,3-Diarylpyrazolyl-Acylsulfonamide Inhibitor

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Mycobacterium Tuberculosis 3-Hydroxyl-Acp Dehydratase Hadab in Complex with 1,3-Diarylpyrazolyl-Acylsulfonamide Inhibitor within 5.0Å range:
|
Reference:
V.Singh,
A.E.Grzegorzewicz,
S.Fienberg,
R.Muller,
L.P.Khonde,
O.Sanz,
S.Alfonso,
B.Urones,
G.Drewes,
M.Bantscheff,
S.Ghidelli-Disse,
T.R.Ioerger,
B.Angala,
J.Liu,
R.E.Lee,
J.C.Sacchettini,
I.V.Krieger,
M.Jackson,
K.Chibale,
S.R.Ghorpade.
1,3-Diarylpyrazolyl-Acylsulfonamides Target Hadab/Bc Complex in Mycobacterium Tuberculosis . Acs Infect Dis. V. 8 2315 2022.
ISSN: ESSN 2373-8227
PubMed: 36325756
DOI: 10.1021/ACSINFECDIS.2C00392
Page generated: Sun Jul 13 07:12:12 2025
ISSN: ESSN 2373-8227
PubMed: 36325756
DOI: 10.1021/ACSINFECDIS.2C00392
Last articles
F in 7QN3F in 7QMY
F in 7QMX
F in 7QMW
F in 7QMZ
F in 7QMU
F in 7QMV
F in 7QMT
F in 7QMS
F in 7QMQ