Chlorine »
PDB 7thh-7tp5 »
7tn4 »
Chlorine in PDB 7tn4: Diphosphoinositol Polyphosphate Phosphohydrolase 1 (DIPP1/NUDT3) in Complex with 3-Diphosphoinositol 1,2,4,5-Tetrakisphosphate (3-Pp- IP4), Mg and Fluoride Ion
Enzymatic activity of Diphosphoinositol Polyphosphate Phosphohydrolase 1 (DIPP1/NUDT3) in Complex with 3-Diphosphoinositol 1,2,4,5-Tetrakisphosphate (3-Pp- IP4), Mg and Fluoride Ion
All present enzymatic activity of Diphosphoinositol Polyphosphate Phosphohydrolase 1 (DIPP1/NUDT3) in Complex with 3-Diphosphoinositol 1,2,4,5-Tetrakisphosphate (3-Pp- IP4), Mg and Fluoride Ion:
3.6.1.52;
3.6.1.52;
Protein crystallography data
The structure of Diphosphoinositol Polyphosphate Phosphohydrolase 1 (DIPP1/NUDT3) in Complex with 3-Diphosphoinositol 1,2,4,5-Tetrakisphosphate (3-Pp- IP4), Mg and Fluoride Ion, PDB code: 7tn4
was solved by
G.Zong,
H.Wang,
S.B.Shears,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 31.40 / 1.85 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 45.149, 59.743, 62.769, 90, 90, 90 |
| R / Rfree (%) | 18.3 / 21.7 |
Other elements in 7tn4:
The structure of Diphosphoinositol Polyphosphate Phosphohydrolase 1 (DIPP1/NUDT3) in Complex with 3-Diphosphoinositol 1,2,4,5-Tetrakisphosphate (3-Pp- IP4), Mg and Fluoride Ion also contains other interesting chemical elements:
| Fluorine | (F) | 1 atom |
| Magnesium | (Mg) | 3 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Diphosphoinositol Polyphosphate Phosphohydrolase 1 (DIPP1/NUDT3) in Complex with 3-Diphosphoinositol 1,2,4,5-Tetrakisphosphate (3-Pp- IP4), Mg and Fluoride Ion
(pdb code 7tn4). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Diphosphoinositol Polyphosphate Phosphohydrolase 1 (DIPP1/NUDT3) in Complex with 3-Diphosphoinositol 1,2,4,5-Tetrakisphosphate (3-Pp- IP4), Mg and Fluoride Ion, PDB code: 7tn4:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Diphosphoinositol Polyphosphate Phosphohydrolase 1 (DIPP1/NUDT3) in Complex with 3-Diphosphoinositol 1,2,4,5-Tetrakisphosphate (3-Pp- IP4), Mg and Fluoride Ion, PDB code: 7tn4:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 7tn4
Go back to
Chlorine binding site 1 out
of 2 in the Diphosphoinositol Polyphosphate Phosphohydrolase 1 (DIPP1/NUDT3) in Complex with 3-Diphosphoinositol 1,2,4,5-Tetrakisphosphate (3-Pp- IP4), Mg and Fluoride Ion
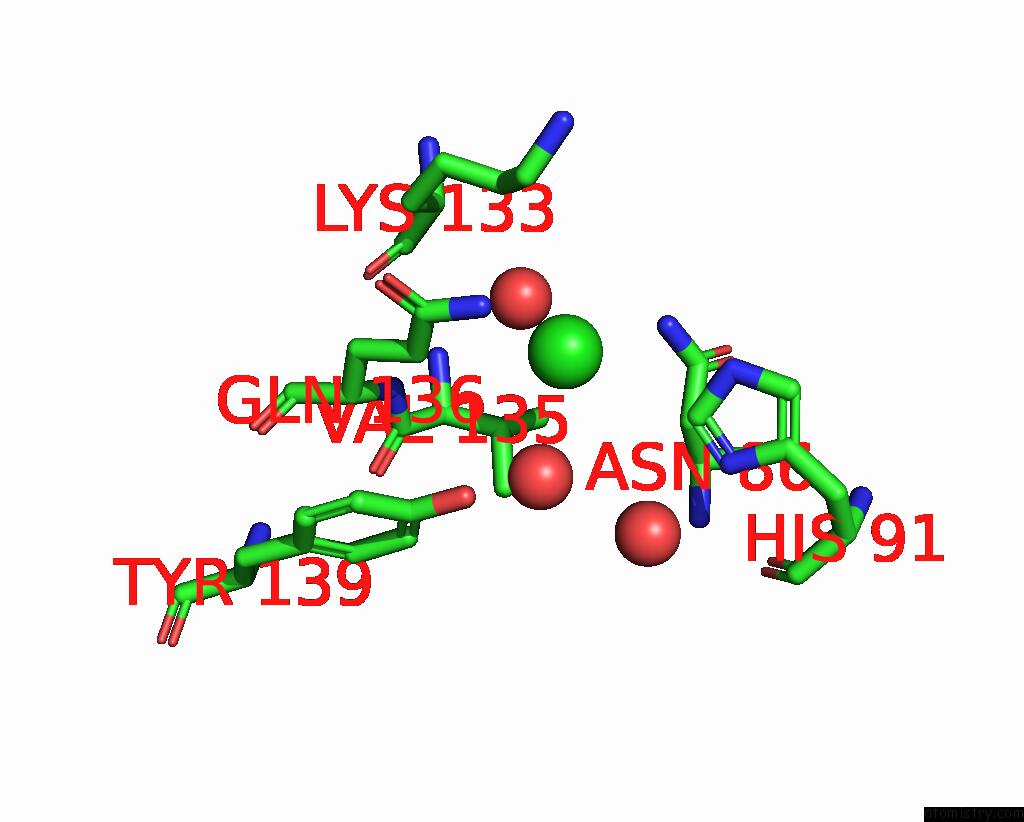
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Diphosphoinositol Polyphosphate Phosphohydrolase 1 (DIPP1/NUDT3) in Complex with 3-Diphosphoinositol 1,2,4,5-Tetrakisphosphate (3-Pp- IP4), Mg and Fluoride Ion within 5.0Å range:
|
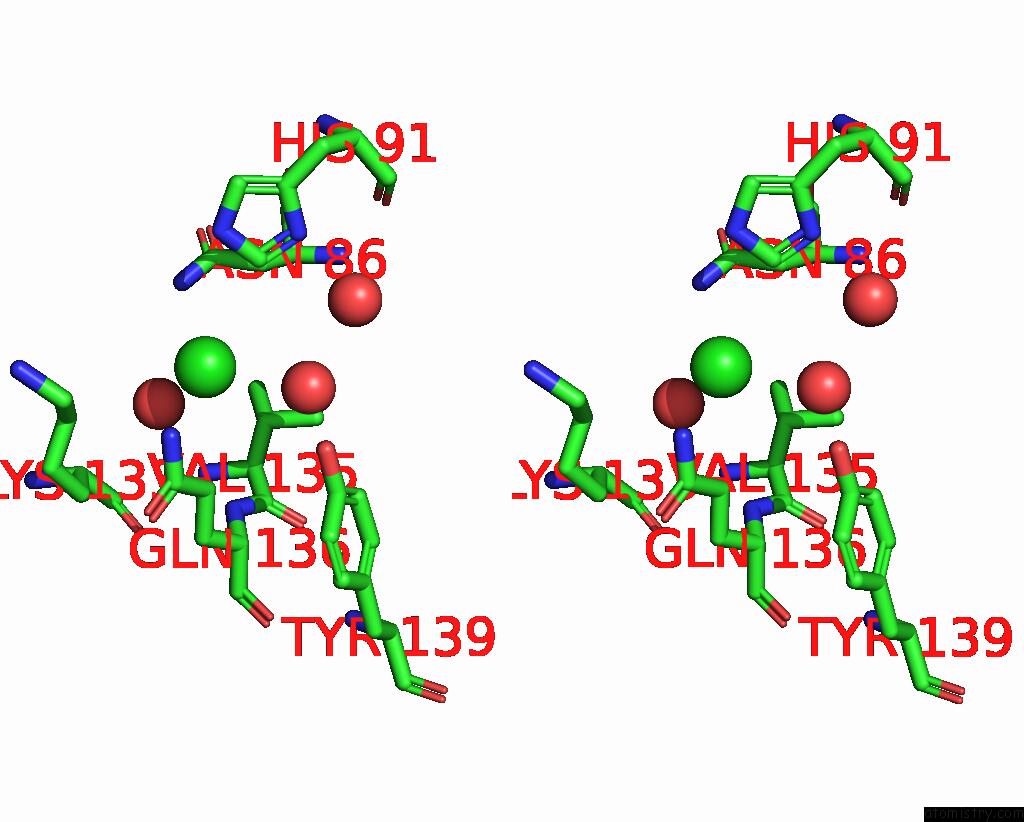
Chlorine binding site 2 out of 2 in 7tn4
Go back to
Chlorine binding site 2 out
of 2 in the Diphosphoinositol Polyphosphate Phosphohydrolase 1 (DIPP1/NUDT3) in Complex with 3-Diphosphoinositol 1,2,4,5-Tetrakisphosphate (3-Pp- IP4), Mg and Fluoride Ion

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Diphosphoinositol Polyphosphate Phosphohydrolase 1 (DIPP1/NUDT3) in Complex with 3-Diphosphoinositol 1,2,4,5-Tetrakisphosphate (3-Pp- IP4), Mg and Fluoride Ion within 5.0Å range:
|
Reference:
G.Zong,
S.B.Shears,
H.Wang.
Structural and Catalytic Analyses of the Insp 6 Kinase Activities of Higher Plant Itpks. Faseb J. V. 36 22380 2022.
ISSN: ESSN 1530-6860
PubMed: 35635723
DOI: 10.1096/FJ.202200393R
Page generated: Sun Jul 13 07:31:20 2025
ISSN: ESSN 1530-6860
PubMed: 35635723
DOI: 10.1096/FJ.202200393R
Last articles
F in 7QMOF in 7QMN
F in 7QMM
F in 7QML
F in 7QMK
F in 7QMJ
F in 7QMI
F in 7QMH
F in 7QMG
F in 7QMF