Chlorine »
PDB 7tp6-7u1d »
7tsp »
Chlorine in PDB 7tsp: Structure of Human Endothelial Nitric Oxide Synthase Heme Domain in Complex with 6-(3-(4,4-Difluoropiperidin-1-Yl)Prop-1-Yn-1-Yl)-4- Methylpyridin-2-Amine
Enzymatic activity of Structure of Human Endothelial Nitric Oxide Synthase Heme Domain in Complex with 6-(3-(4,4-Difluoropiperidin-1-Yl)Prop-1-Yn-1-Yl)-4- Methylpyridin-2-Amine
All present enzymatic activity of Structure of Human Endothelial Nitric Oxide Synthase Heme Domain in Complex with 6-(3-(4,4-Difluoropiperidin-1-Yl)Prop-1-Yn-1-Yl)-4- Methylpyridin-2-Amine:
1.14.13.39;
1.14.13.39;
Protein crystallography data
The structure of Structure of Human Endothelial Nitric Oxide Synthase Heme Domain in Complex with 6-(3-(4,4-Difluoropiperidin-1-Yl)Prop-1-Yn-1-Yl)-4- Methylpyridin-2-Amine, PDB code: 7tsp
was solved by
H.Li,
T.L.Poulos,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 39.02 / 2.00 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 59.471, 152.71, 108.781, 90, 90.62, 90 |
| R / Rfree (%) | 21.5 / 26.7 |
Other elements in 7tsp:
The structure of Structure of Human Endothelial Nitric Oxide Synthase Heme Domain in Complex with 6-(3-(4,4-Difluoropiperidin-1-Yl)Prop-1-Yn-1-Yl)-4- Methylpyridin-2-Amine also contains other interesting chemical elements:
| Fluorine | (F) | 8 atoms |
| Zinc | (Zn) | 2 atoms |
| Iron | (Fe) | 4 atoms |
| Gadolinium | (Gd) | 4 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Structure of Human Endothelial Nitric Oxide Synthase Heme Domain in Complex with 6-(3-(4,4-Difluoropiperidin-1-Yl)Prop-1-Yn-1-Yl)-4- Methylpyridin-2-Amine
(pdb code 7tsp). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 4 binding sites of Chlorine where determined in the Structure of Human Endothelial Nitric Oxide Synthase Heme Domain in Complex with 6-(3-(4,4-Difluoropiperidin-1-Yl)Prop-1-Yn-1-Yl)-4- Methylpyridin-2-Amine, PDB code: 7tsp:
Jump to Chlorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Chlorine where determined in the Structure of Human Endothelial Nitric Oxide Synthase Heme Domain in Complex with 6-(3-(4,4-Difluoropiperidin-1-Yl)Prop-1-Yn-1-Yl)-4- Methylpyridin-2-Amine, PDB code: 7tsp:
Jump to Chlorine binding site number: 1; 2; 3; 4;
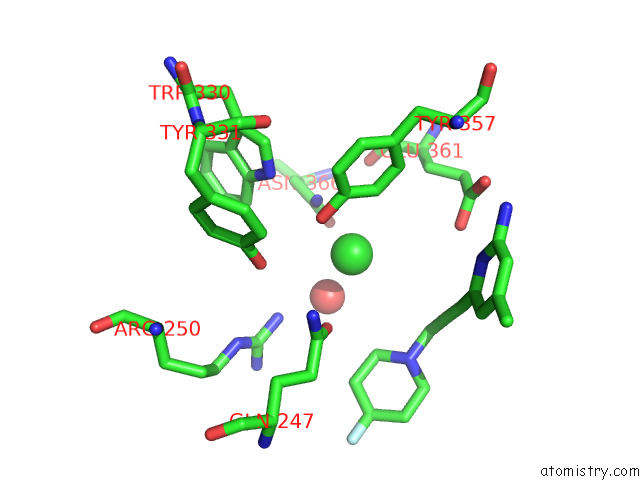
Chlorine binding site 1 out of 4 in 7tsp
Go back to
Chlorine binding site 1 out
of 4 in the Structure of Human Endothelial Nitric Oxide Synthase Heme Domain in Complex with 6-(3-(4,4-Difluoropiperidin-1-Yl)Prop-1-Yn-1-Yl)-4- Methylpyridin-2-Amine
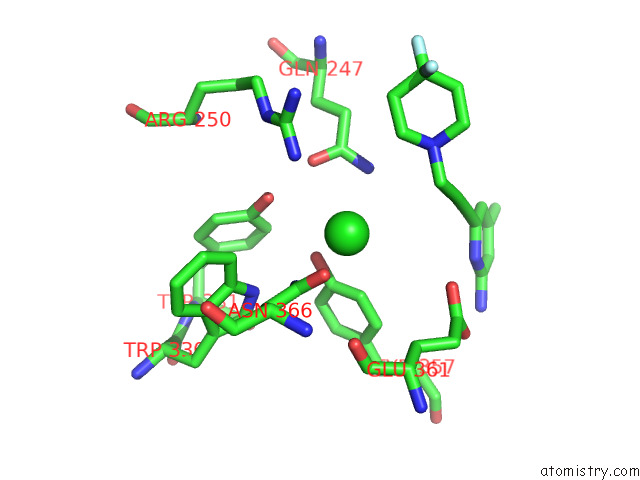
Mono view
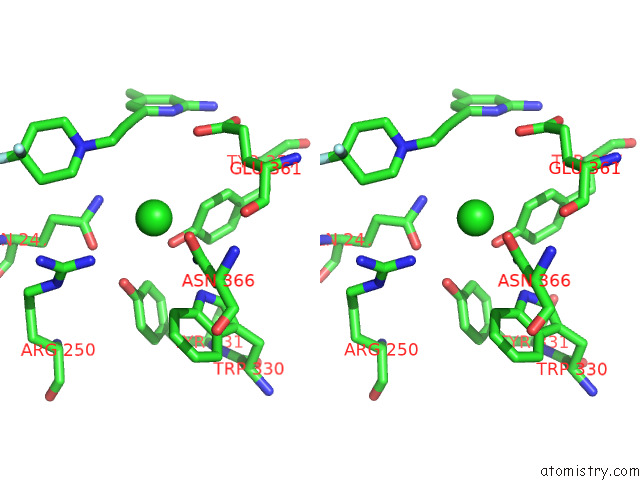
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Structure of Human Endothelial Nitric Oxide Synthase Heme Domain in Complex with 6-(3-(4,4-Difluoropiperidin-1-Yl)Prop-1-Yn-1-Yl)-4- Methylpyridin-2-Amine within 5.0Å range:
|
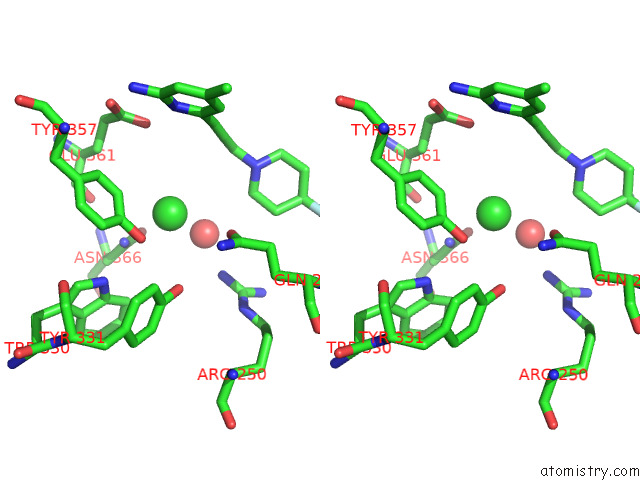
Chlorine binding site 2 out of 4 in 7tsp
Go back to
Chlorine binding site 2 out
of 4 in the Structure of Human Endothelial Nitric Oxide Synthase Heme Domain in Complex with 6-(3-(4,4-Difluoropiperidin-1-Yl)Prop-1-Yn-1-Yl)-4- Methylpyridin-2-Amine
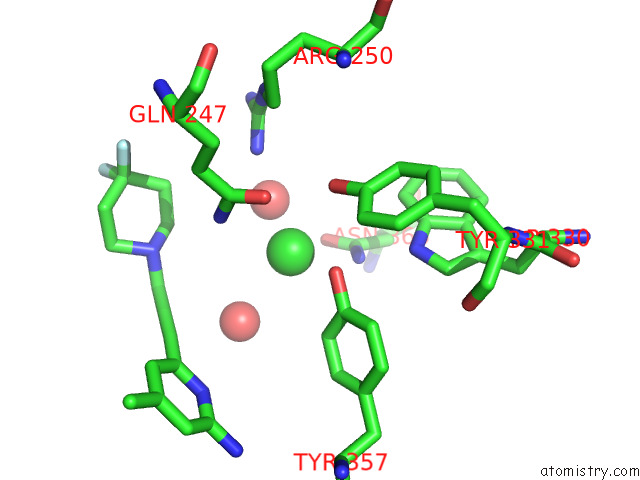
Mono view
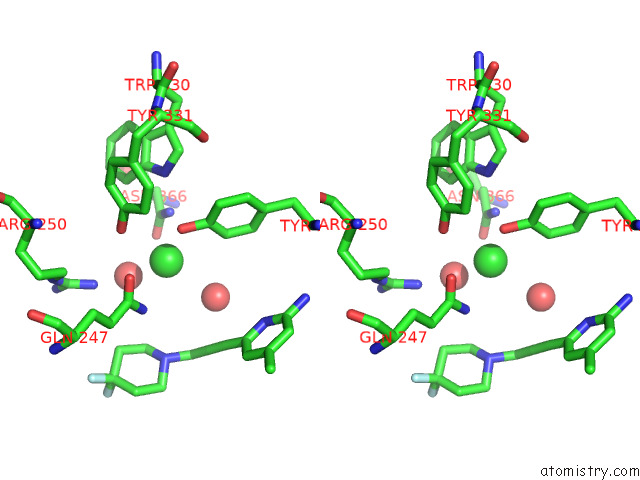
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Structure of Human Endothelial Nitric Oxide Synthase Heme Domain in Complex with 6-(3-(4,4-Difluoropiperidin-1-Yl)Prop-1-Yn-1-Yl)-4- Methylpyridin-2-Amine within 5.0Å range:
|
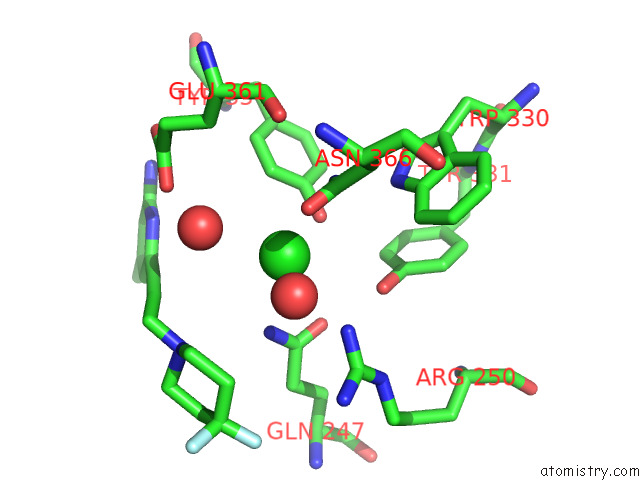
Chlorine binding site 3 out of 4 in 7tsp
Go back to
Chlorine binding site 3 out
of 4 in the Structure of Human Endothelial Nitric Oxide Synthase Heme Domain in Complex with 6-(3-(4,4-Difluoropiperidin-1-Yl)Prop-1-Yn-1-Yl)-4- Methylpyridin-2-Amine
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Structure of Human Endothelial Nitric Oxide Synthase Heme Domain in Complex with 6-(3-(4,4-Difluoropiperidin-1-Yl)Prop-1-Yn-1-Yl)-4- Methylpyridin-2-Amine within 5.0Å range:
|
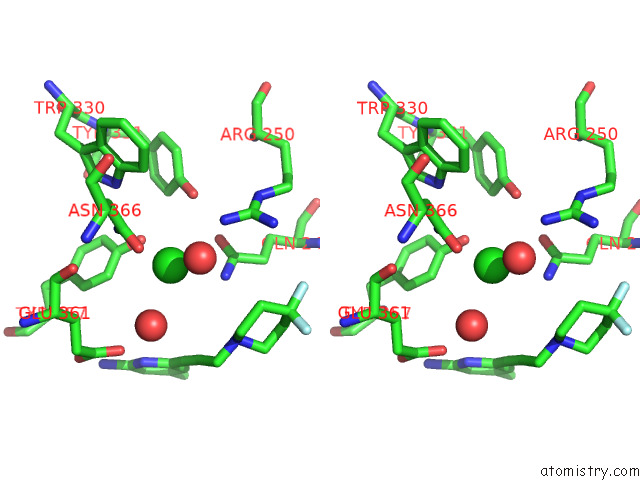
Chlorine binding site 4 out of 4 in 7tsp
Go back to
Chlorine binding site 4 out
of 4 in the Structure of Human Endothelial Nitric Oxide Synthase Heme Domain in Complex with 6-(3-(4,4-Difluoropiperidin-1-Yl)Prop-1-Yn-1-Yl)-4- Methylpyridin-2-Amine
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Structure of Human Endothelial Nitric Oxide Synthase Heme Domain in Complex with 6-(3-(4,4-Difluoropiperidin-1-Yl)Prop-1-Yn-1-Yl)-4- Methylpyridin-2-Amine within 5.0Å range:
|
Reference:
D.Vasu,
H.Li,
C.D.Hardy,
T.L.Poulos,
R.B.Silverman.
2-Aminopyridines with A Shortened Amino Sidechain As Potent, Selective, and Highly Permeable Human Neuronal Nitric Oxide Synthase Inhibitors. Bioorg.Med.Chem. V. 69 16878 2022.
ISSN: ESSN 1464-3391
PubMed: 35772285
DOI: 10.1016/J.BMC.2022.116878
Page generated: Sun Jul 13 07:38:29 2025
ISSN: ESSN 1464-3391
PubMed: 35772285
DOI: 10.1016/J.BMC.2022.116878
Last articles
F in 7PRMF in 7PQV
F in 7PQS
F in 7PMM
F in 7POL
F in 7POR
F in 7PNS
F in 7P81
F in 7PLI
F in 7PJR