Chlorine »
PDB 8a19-8aau »
8a6g »
Chlorine in PDB 8a6g: Room Temperature RSEGFP2 with A Chlorinated Chromophore in the Non- Fluorescent Off-State
Protein crystallography data
The structure of Room Temperature RSEGFP2 with A Chlorinated Chromophore in the Non- Fluorescent Off-State, PDB code: 8a6g
was solved by
A.Fadini,
J.Van Thor,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 31.46 / 1.63 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 51.99, 62.91, 72.03, 90, 90, 90 |
| R / Rfree (%) | 15.5 / 19.5 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Room Temperature RSEGFP2 with A Chlorinated Chromophore in the Non- Fluorescent Off-State
(pdb code 8a6g). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 3 binding sites of Chlorine where determined in the Room Temperature RSEGFP2 with A Chlorinated Chromophore in the Non- Fluorescent Off-State, PDB code: 8a6g:
Jump to Chlorine binding site number: 1; 2; 3;
In total 3 binding sites of Chlorine where determined in the Room Temperature RSEGFP2 with A Chlorinated Chromophore in the Non- Fluorescent Off-State, PDB code: 8a6g:
Jump to Chlorine binding site number: 1; 2; 3;
Chlorine binding site 1 out of 3 in 8a6g
Go back to
Chlorine binding site 1 out
of 3 in the Room Temperature RSEGFP2 with A Chlorinated Chromophore in the Non- Fluorescent Off-State
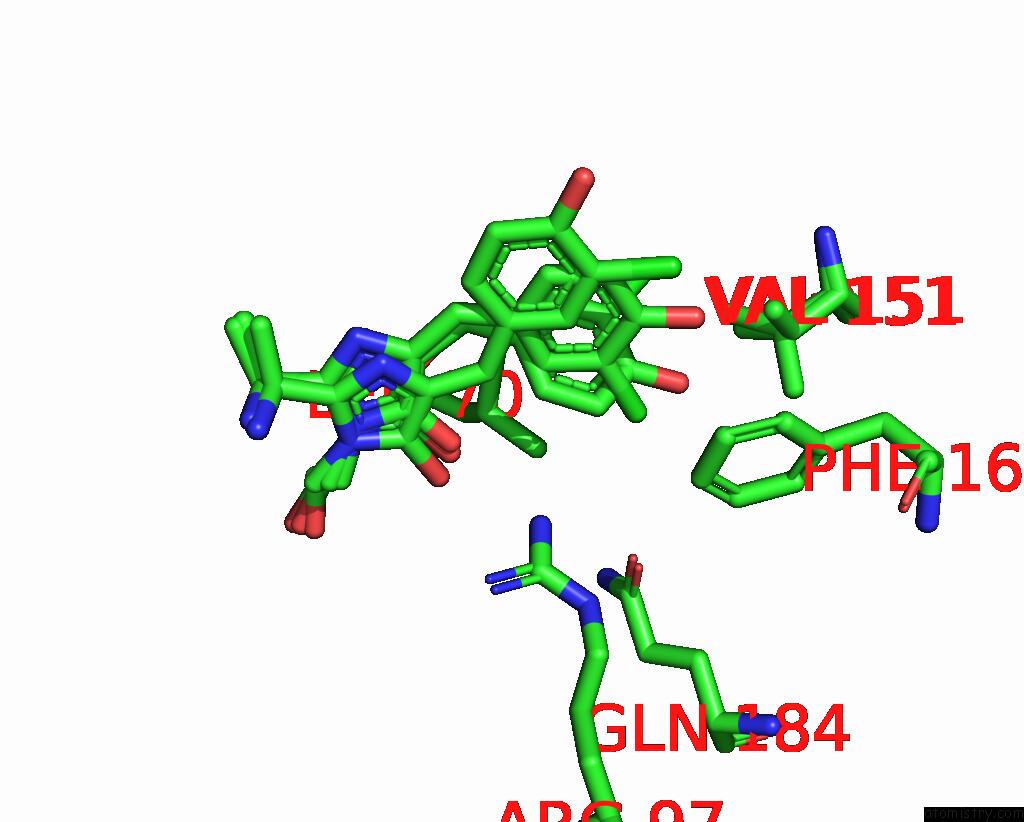
Mono view
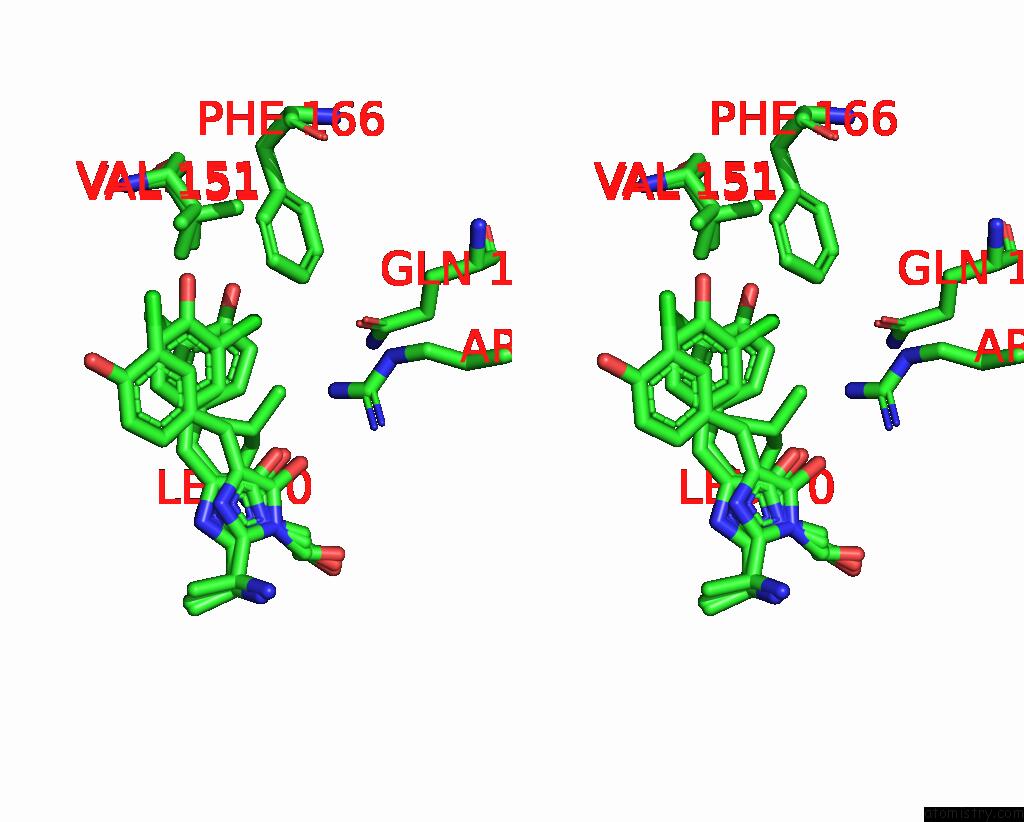
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Room Temperature RSEGFP2 with A Chlorinated Chromophore in the Non- Fluorescent Off-State within 5.0Å range:
|
Chlorine binding site 2 out of 3 in 8a6g
Go back to
Chlorine binding site 2 out
of 3 in the Room Temperature RSEGFP2 with A Chlorinated Chromophore in the Non- Fluorescent Off-State
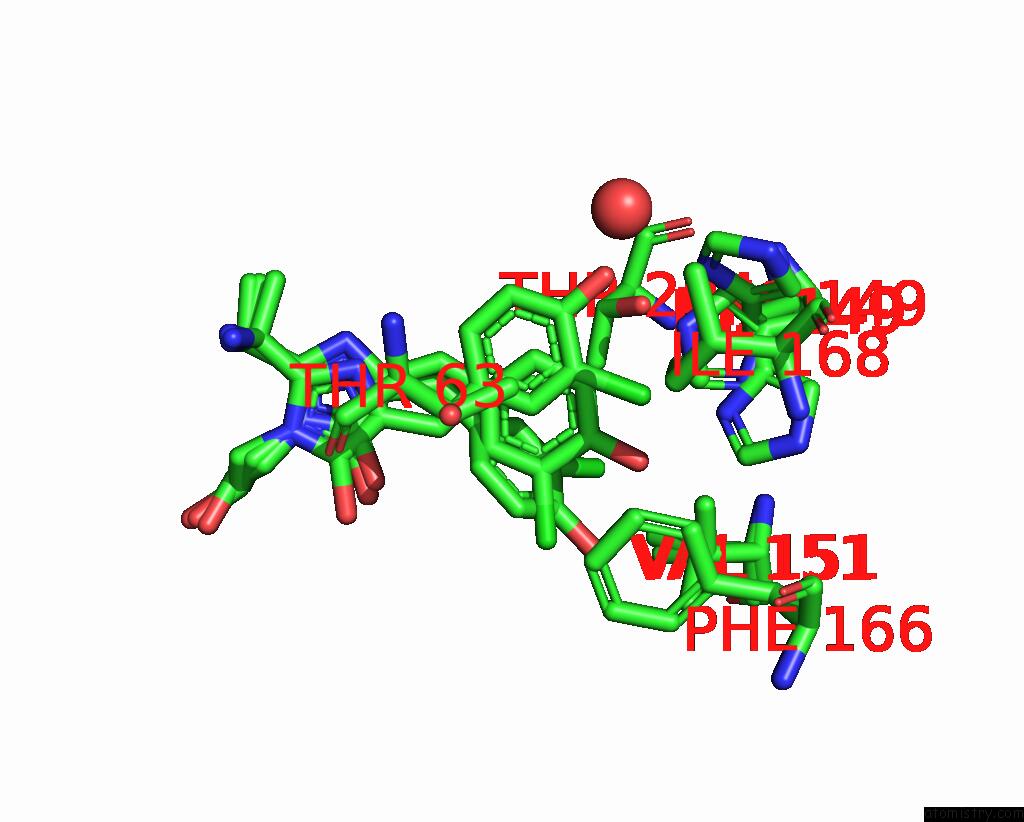
Mono view
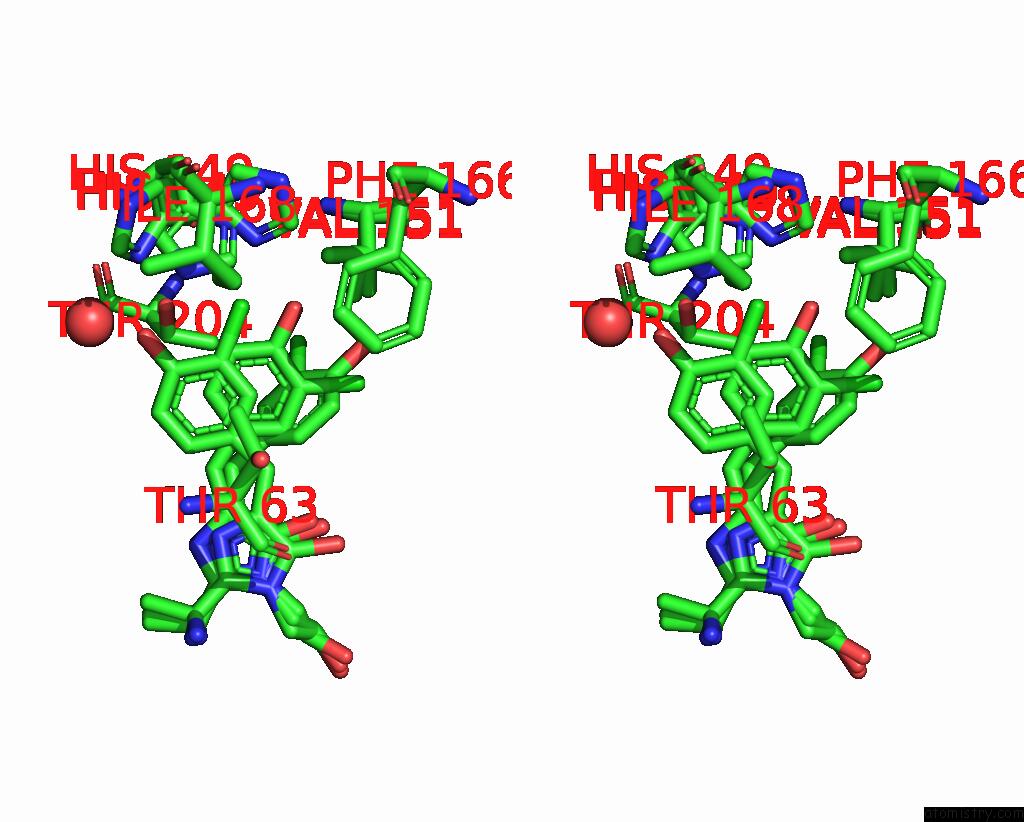
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Room Temperature RSEGFP2 with A Chlorinated Chromophore in the Non- Fluorescent Off-State within 5.0Å range:
|
Chlorine binding site 3 out of 3 in 8a6g
Go back to
Chlorine binding site 3 out
of 3 in the Room Temperature RSEGFP2 with A Chlorinated Chromophore in the Non- Fluorescent Off-State

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Room Temperature RSEGFP2 with A Chlorinated Chromophore in the Non- Fluorescent Off-State within 5.0Å range:
|
Reference:
A.Fadini,
C.D.M.Hutchison,
D.Morozov,
J.Chang,
K.Maghlaoui,
S.Perrett,
F.Luo,
J.C.X.Kho,
M.G.Romei,
R.M.L.Morgan,
C.M.Orr,
V.Cordon-Preciado,
T.Fujiwara,
N.Nuemket,
T.Tosha,
R.Tanaka,
S.Owada,
K.Tono,
S.Iwata,
S.G.Boxer,
G.Groenhof,
E.Nango,
J.J.Van Thor.
Serial Femtosecond Crystallography Reveals That Photoactivation in A Fluorescent Protein Proceeds Via the Hula Twist Mechanism. J.Am.Chem.Soc. 2023.
ISSN: ESSN 1520-5126
PubMed: 37418747
DOI: 10.1021/JACS.3C02313
Page generated: Sun Jul 13 08:59:51 2025
ISSN: ESSN 1520-5126
PubMed: 37418747
DOI: 10.1021/JACS.3C02313
Last articles
F in 4G1WF in 4G16
F in 4FXY
F in 4FXQ
F in 4FZF
F in 4FP1
F in 4FVX
F in 4FV1
F in 4FV9
F in 4FV3