Chlorine »
PDB 8ayq-8b7b »
8b56 »
Chlorine in PDB 8b56: Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9
Enzymatic activity of Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9
All present enzymatic activity of Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9:
3.4.22.69;
3.4.22.69;
Protein crystallography data
The structure of Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9, PDB code: 8b56
was solved by
N.Straeter,
C.Mueller,
T.Claff,
K.Sylvester,
R.Weisse,
S.Gao,
L.Song,
X.Liu,
P.Zhan,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 56.60 / 1.82 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 67.833, 99.107, 102.706, 90, 90, 90 |
| R / Rfree (%) | 18.5 / 23.1 |
Other elements in 8b56:
The structure of Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9 also contains other interesting chemical elements:
| Bromine | (Br) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9
(pdb code 8b56). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 10 binding sites of Chlorine where determined in the Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9, PDB code: 8b56:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
In total 10 binding sites of Chlorine where determined in the Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9, PDB code: 8b56:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Chlorine binding site 1 out of 10 in 8b56
Go back to
Chlorine binding site 1 out
of 10 in the Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9
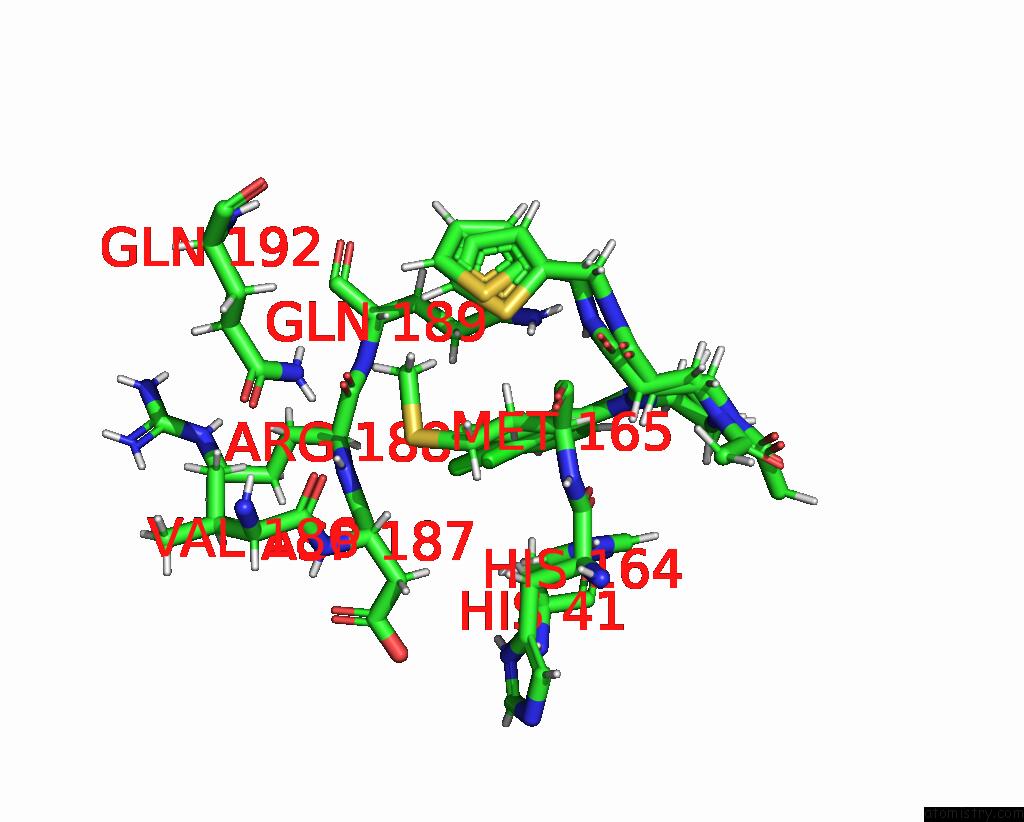
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9 within 5.0Å range:
|
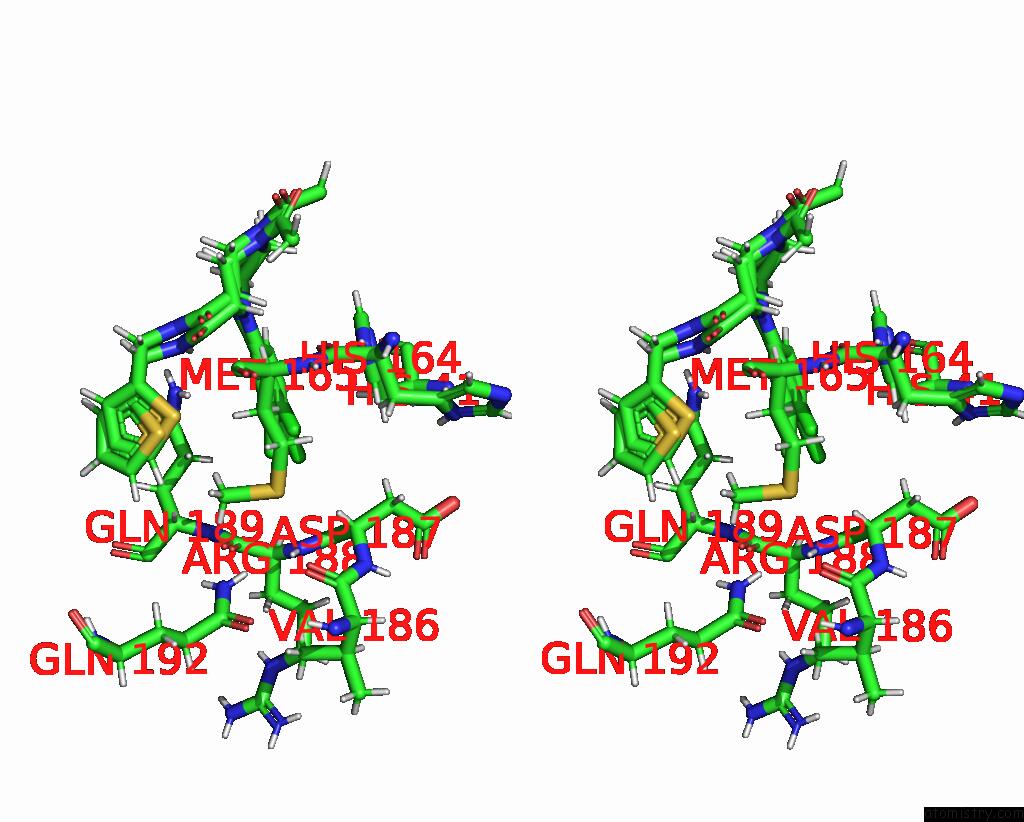
Chlorine binding site 2 out of 10 in 8b56
Go back to
Chlorine binding site 2 out
of 10 in the Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9 within 5.0Å range:
|
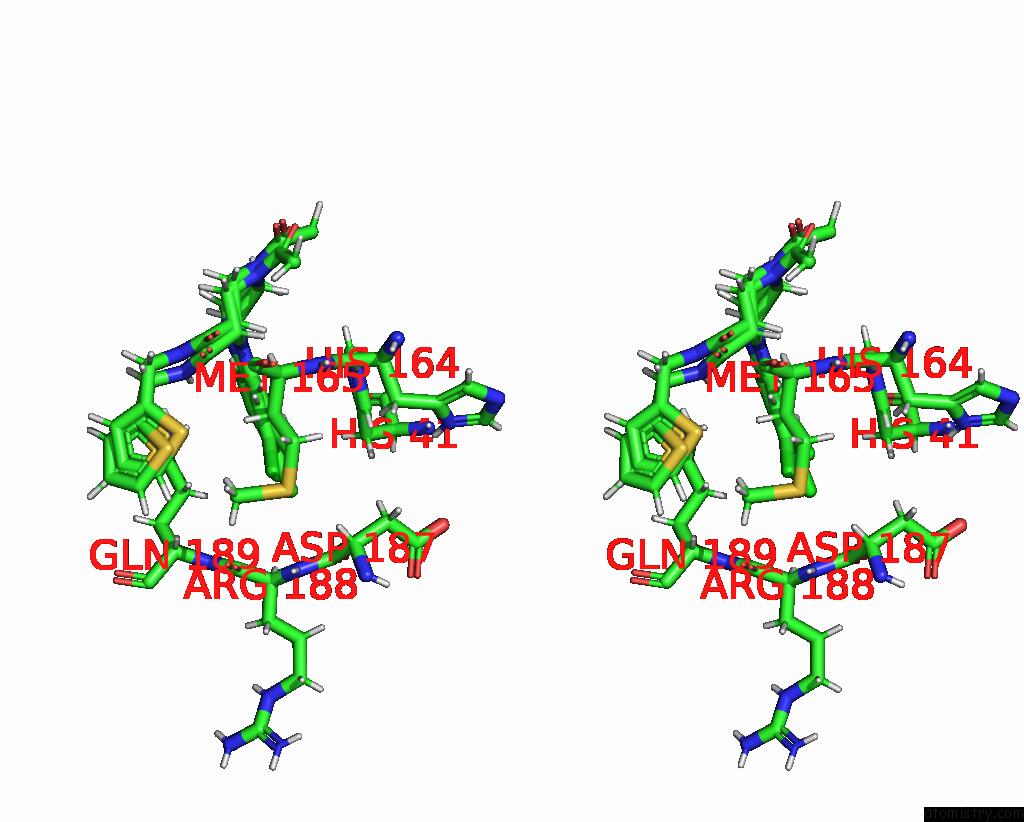
Chlorine binding site 3 out of 10 in 8b56
Go back to
Chlorine binding site 3 out
of 10 in the Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9 within 5.0Å range:
|
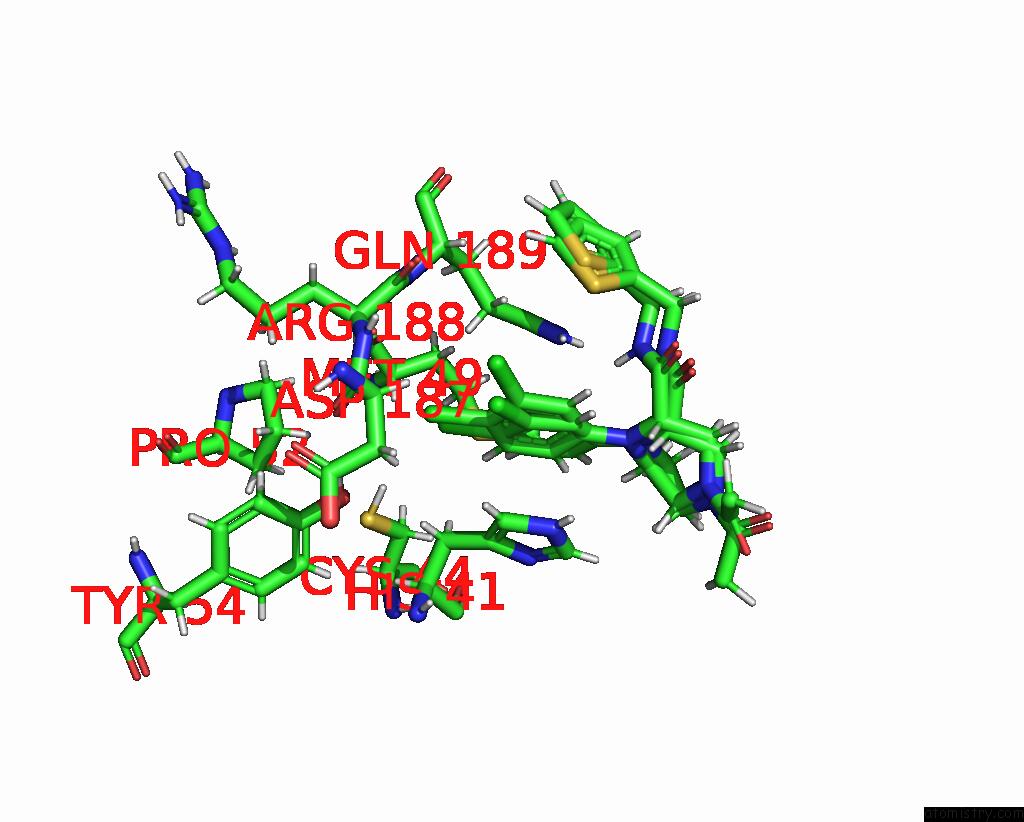
Chlorine binding site 4 out of 10 in 8b56
Go back to
Chlorine binding site 4 out
of 10 in the Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9 within 5.0Å range:
|
Chlorine binding site 5 out of 10 in 8b56
Go back to
Chlorine binding site 5 out
of 10 in the Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9 within 5.0Å range:
|
Chlorine binding site 6 out of 10 in 8b56
Go back to
Chlorine binding site 6 out
of 10 in the Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9
Mono view
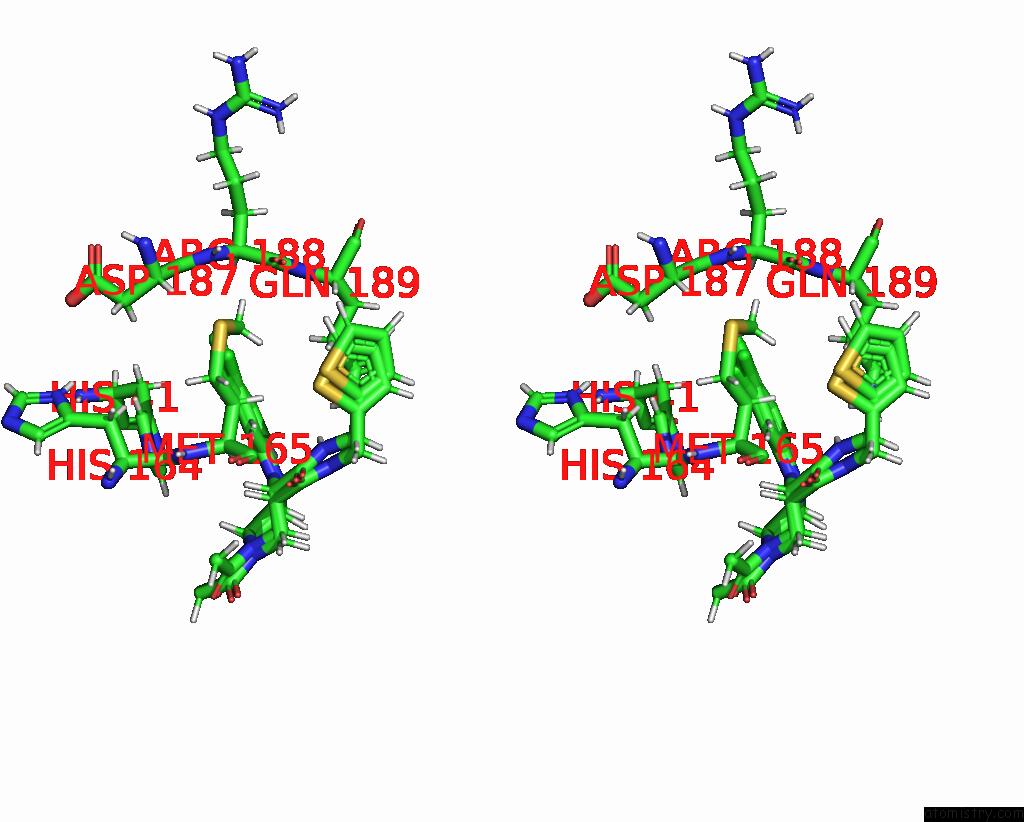
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9 within 5.0Å range:
|
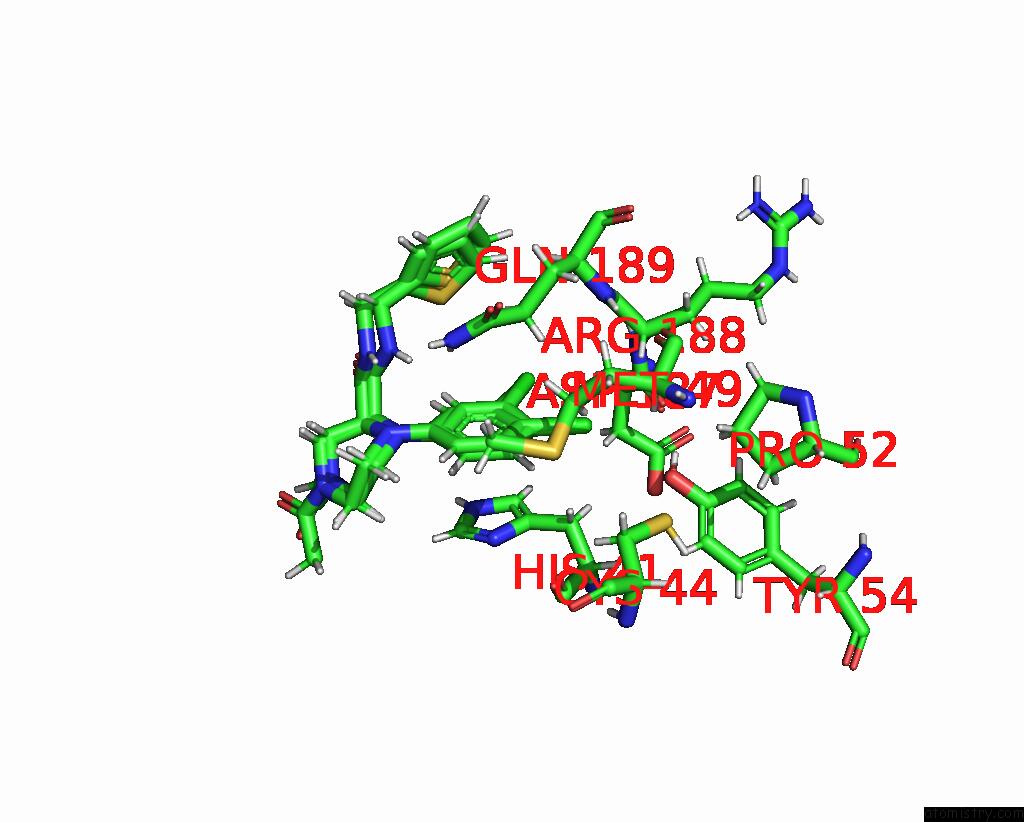
Chlorine binding site 7 out of 10 in 8b56
Go back to
Chlorine binding site 7 out
of 10 in the Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9
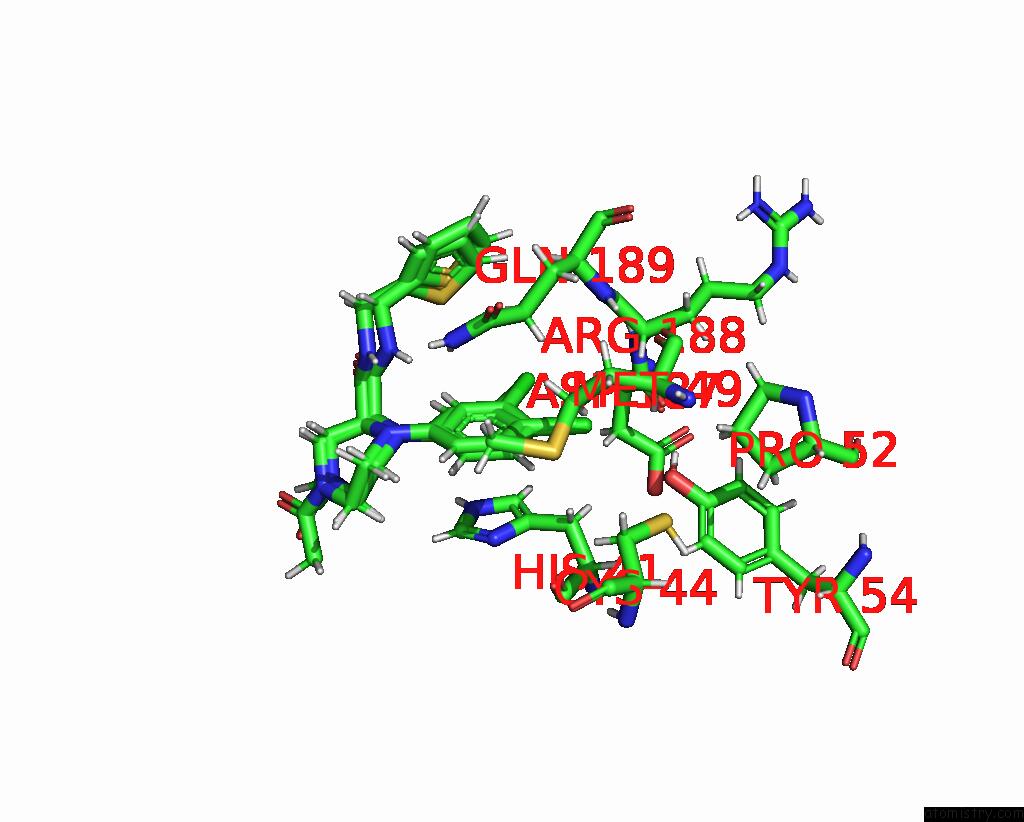
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 7 of Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9 within 5.0Å range:
|
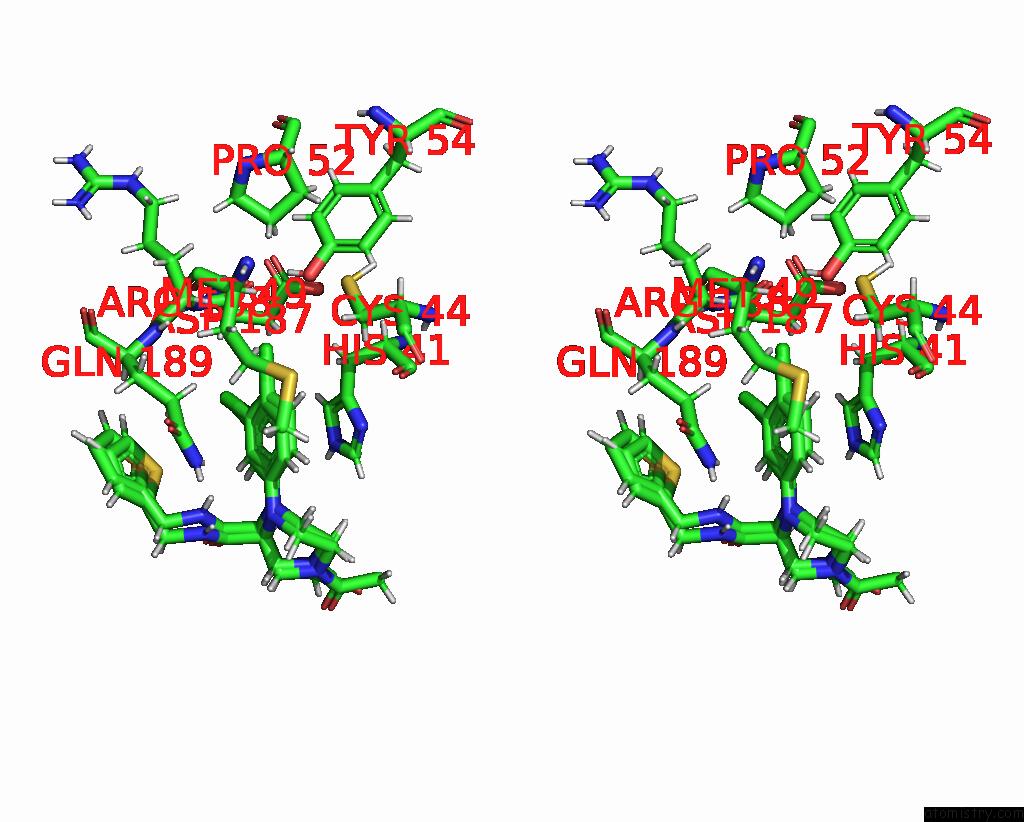
Chlorine binding site 8 out of 10 in 8b56
Go back to
Chlorine binding site 8 out
of 10 in the Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 8 of Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9 within 5.0Å range:
|
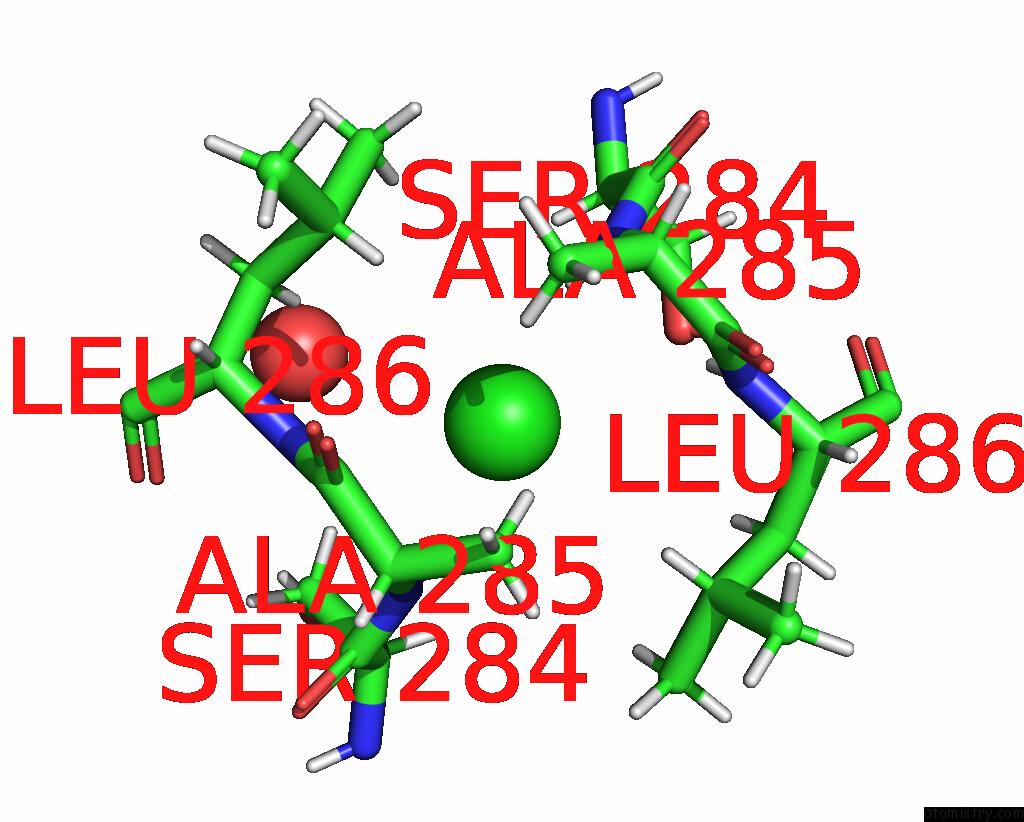
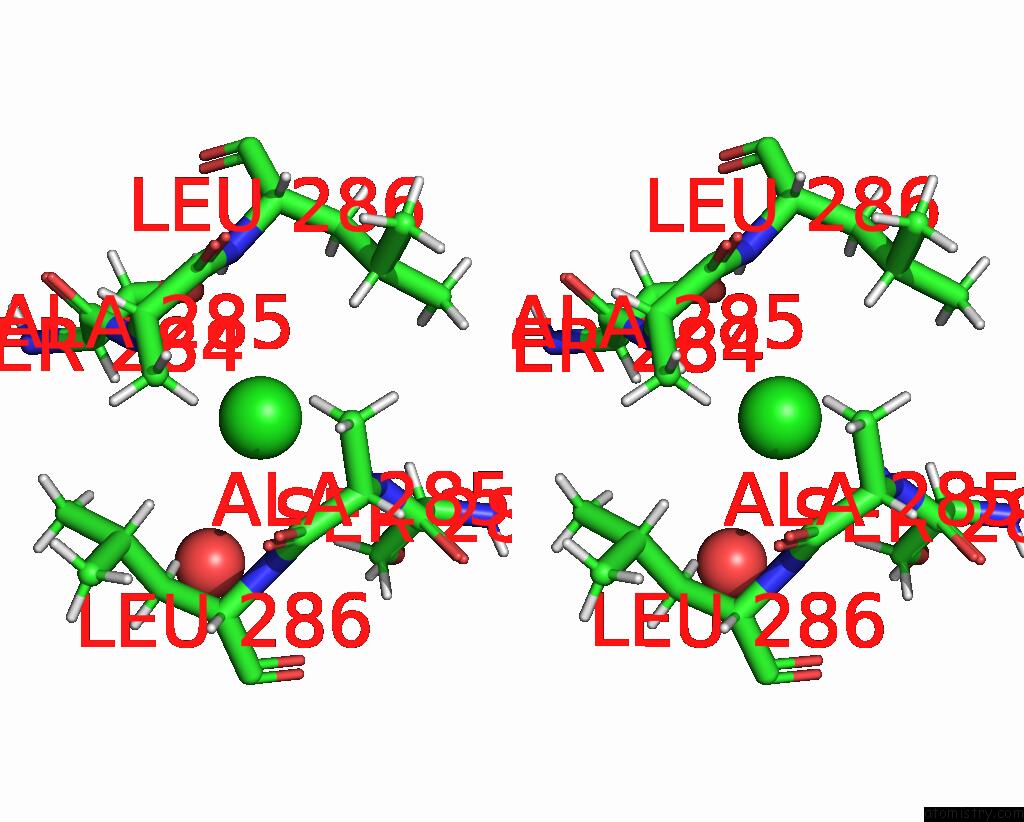
Chlorine binding site 9 out of 10 in 8b56
Go back to
Chlorine binding site 9 out
of 10 in the Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 9 of Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9 within 5.0Å range:
|
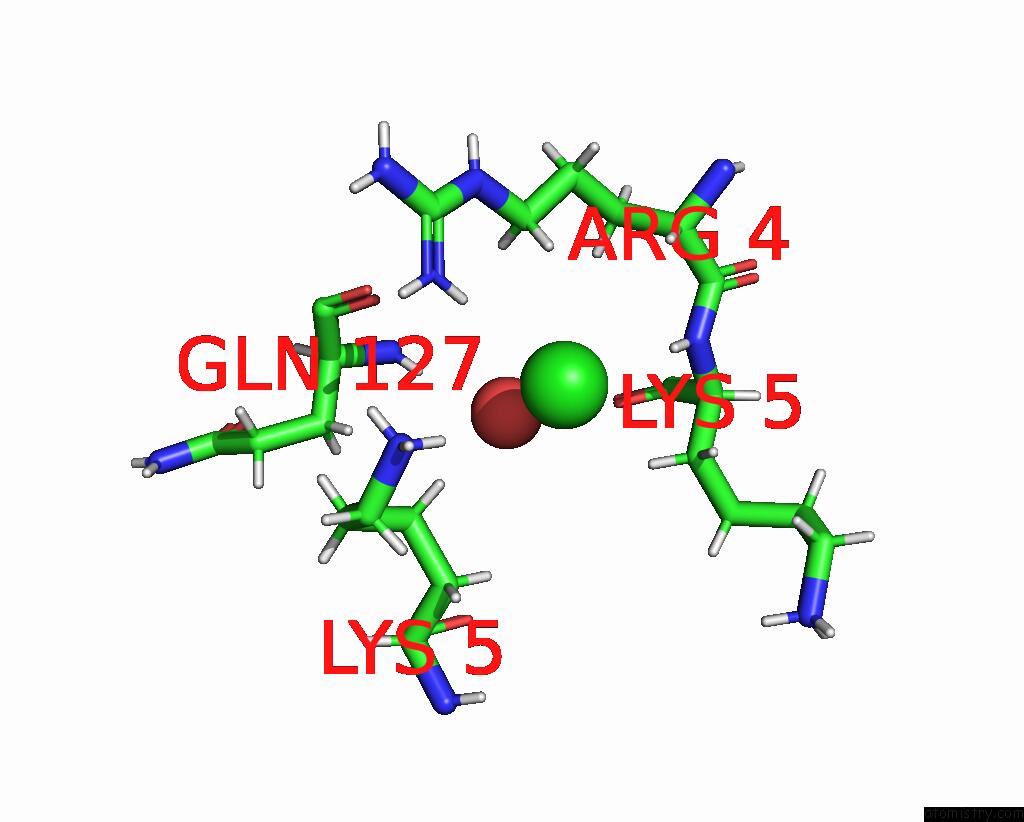
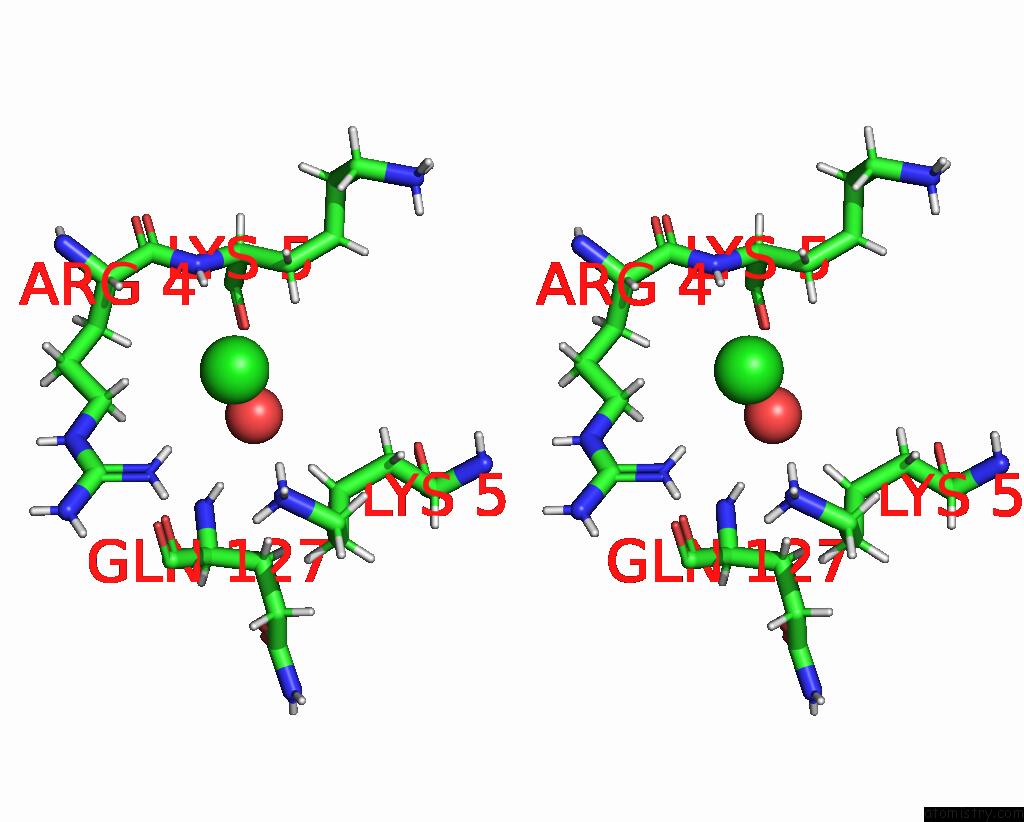
Chlorine binding site 10 out of 10 in 8b56
Go back to
Chlorine binding site 10 out
of 10 in the Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 10 of Crystal Structure of Sars-Cov-2 Main Protease (Mpro) in Complex with the Inhibitor Gd-9 within 5.0Å range:
|
Reference:
S.Gao,
L.Song,
T.Claff,
M.Woodson,
K.Sylvester,
L.Jing,
R.H.Weisse,
Y.Cheng,
N.Strater,
L.Schakel,
M.Gutschow,
B.Ye,
M.Yang,
T.Zhang,
D.Kang,
K.Toth,
J.Tavis,
A.E.Tollefson,
C.E.Muller,
P.Zhan,
X.Liu.
Discovery and Crystallographic Studies of Nonpeptidic Piperazine Derivatives As Covalent Sars-Cov-2 Main Protease Inhibitors. J.Med.Chem. V. 65 16902 2022.
ISSN: ISSN 0022-2623
PubMed: 36475694
DOI: 10.1021/ACS.JMEDCHEM.2C01716
Page generated: Sun Jul 13 09:25:34 2025
ISSN: ISSN 0022-2623
PubMed: 36475694
DOI: 10.1021/ACS.JMEDCHEM.2C01716
Last articles
F in 4GHTF in 4GJ2
F in 4GHI
F in 4GG7
F in 4GG5
F in 4GE6
F in 4GE5
F in 4GCA
F in 4GE2
F in 4G9C