Chlorine »
PDB 8ckq-8ctm »
8cnz »
Chlorine in PDB 8cnz: Mmlare-[4FE-4S] Phased By Fe-Sad
Protein crystallography data
The structure of Mmlare-[4FE-4S] Phased By Fe-Sad, PDB code: 8cnz
was solved by
L.Pecqueur,
P.Zecchin,
B.Golinelli-Pimpaneau,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 55.37 / 3.60 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 187.516, 209.215, 195.799, 90, 90, 90 |
| R / Rfree (%) | 25.3 / 28 |
Other elements in 8cnz:
The structure of Mmlare-[4FE-4S] Phased By Fe-Sad also contains other interesting chemical elements:
| Iron | (Fe) | 28 atoms |
| Iodine | (I) | 4 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Mmlare-[4FE-4S] Phased By Fe-Sad
(pdb code 8cnz). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 5 binding sites of Chlorine where determined in the Mmlare-[4FE-4S] Phased By Fe-Sad, PDB code: 8cnz:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Chlorine where determined in the Mmlare-[4FE-4S] Phased By Fe-Sad, PDB code: 8cnz:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5;
Chlorine binding site 1 out of 5 in 8cnz
Go back to
Chlorine binding site 1 out
of 5 in the Mmlare-[4FE-4S] Phased By Fe-Sad
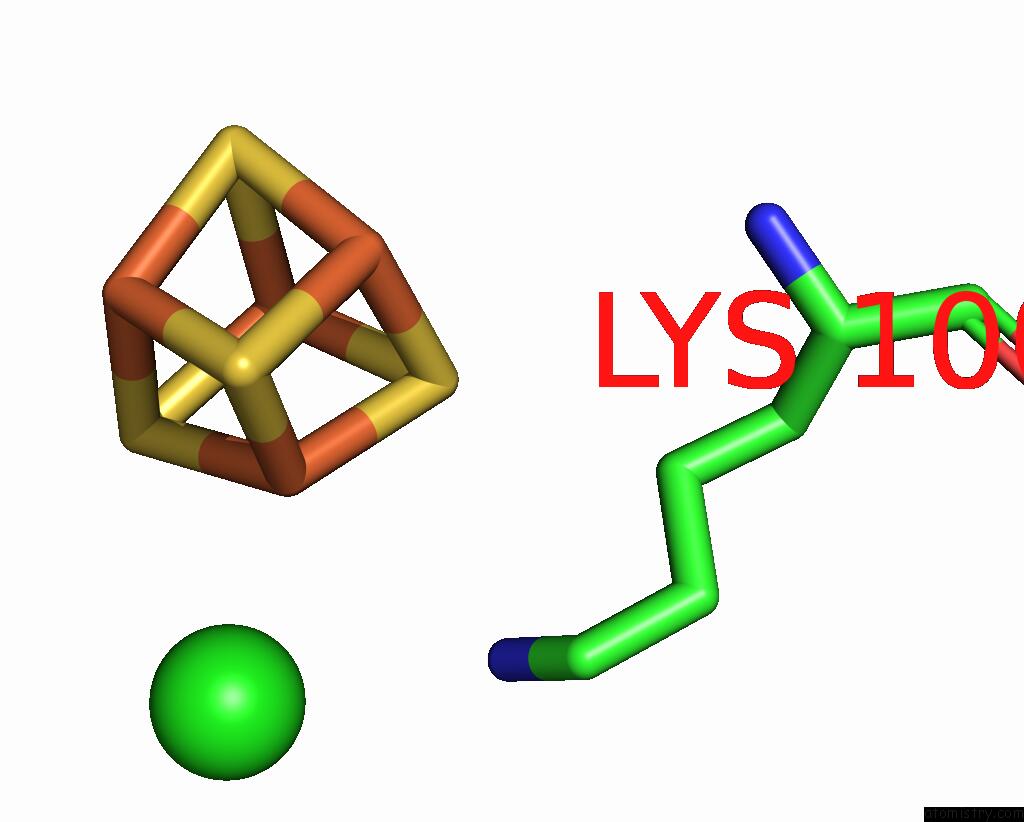
Mono view
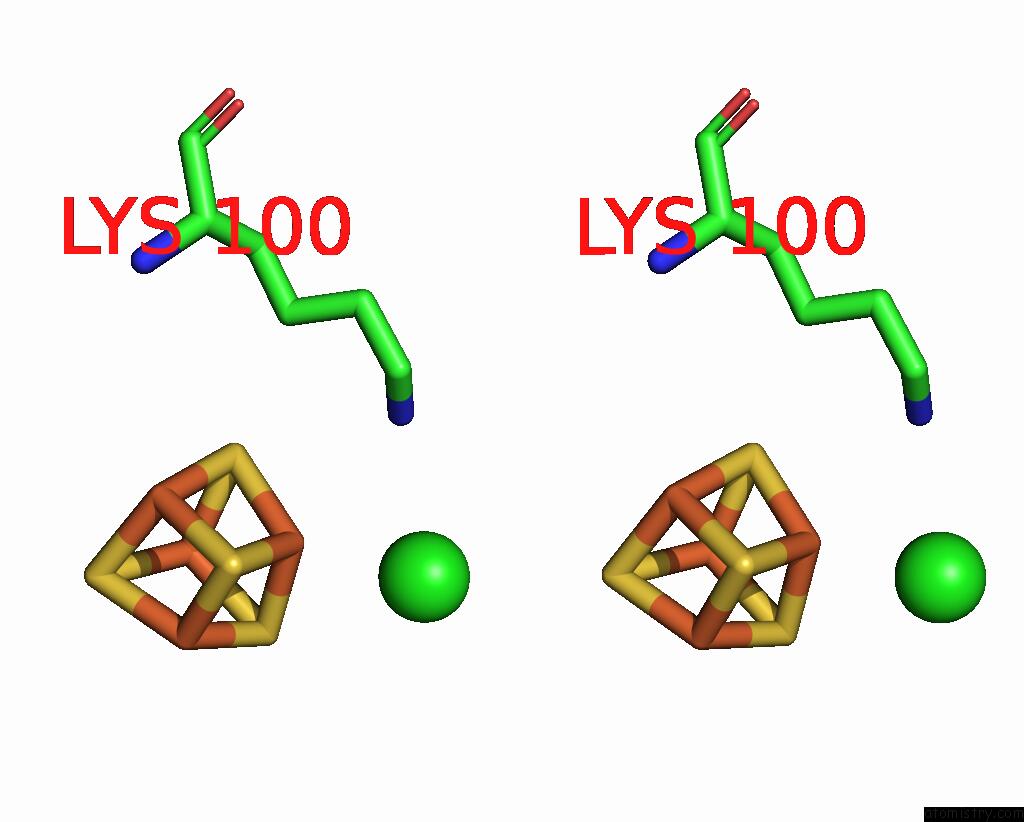
Stereo pair view
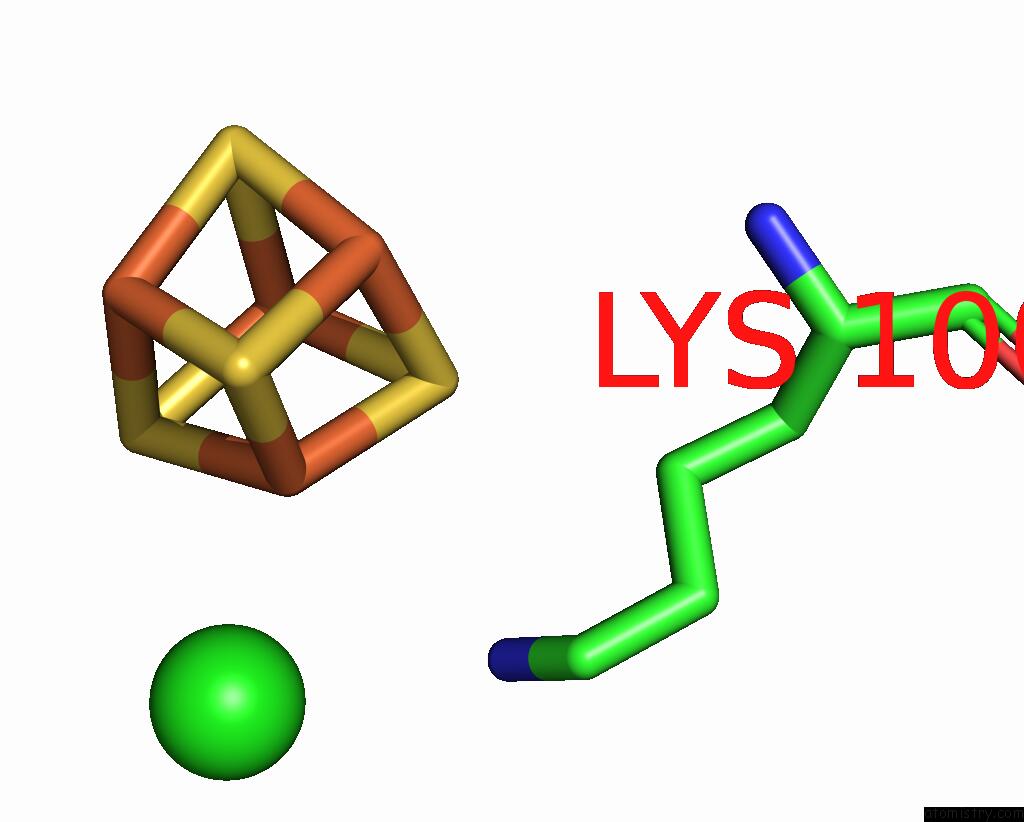
Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Mmlare-[4FE-4S] Phased By Fe-Sad within 5.0Å range:
|
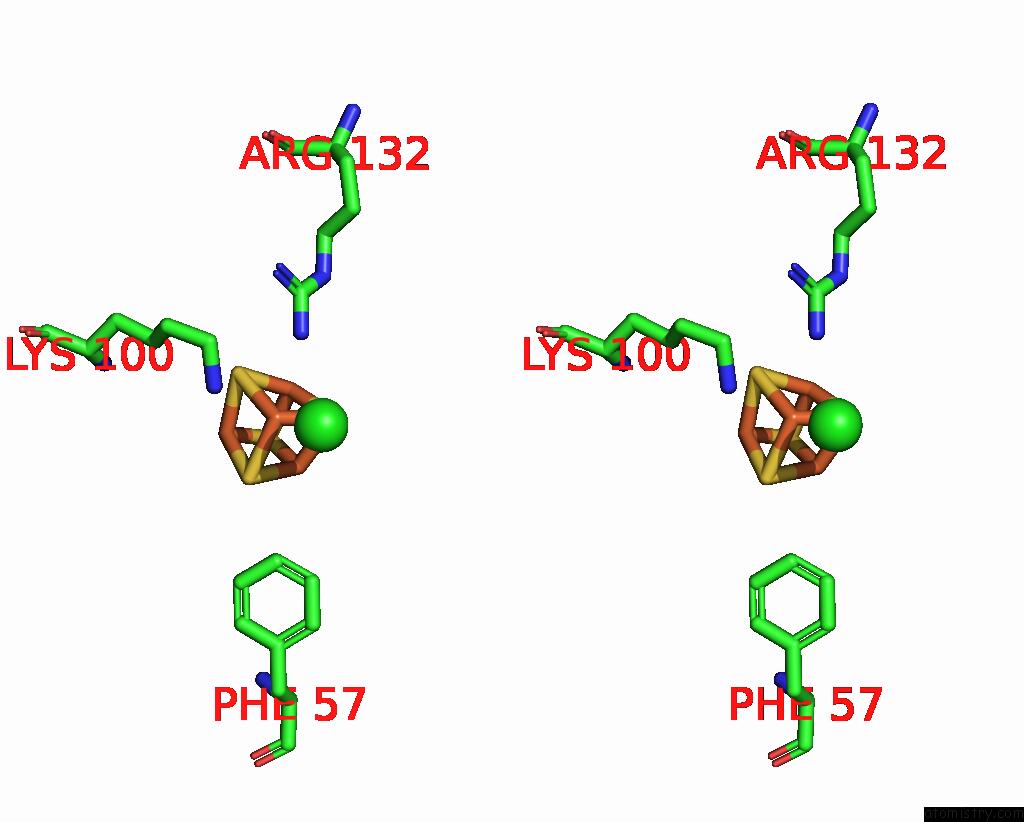
Chlorine binding site 2 out of 5 in 8cnz
Go back to
Chlorine binding site 2 out
of 5 in the Mmlare-[4FE-4S] Phased By Fe-Sad
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Mmlare-[4FE-4S] Phased By Fe-Sad within 5.0Å range:
|
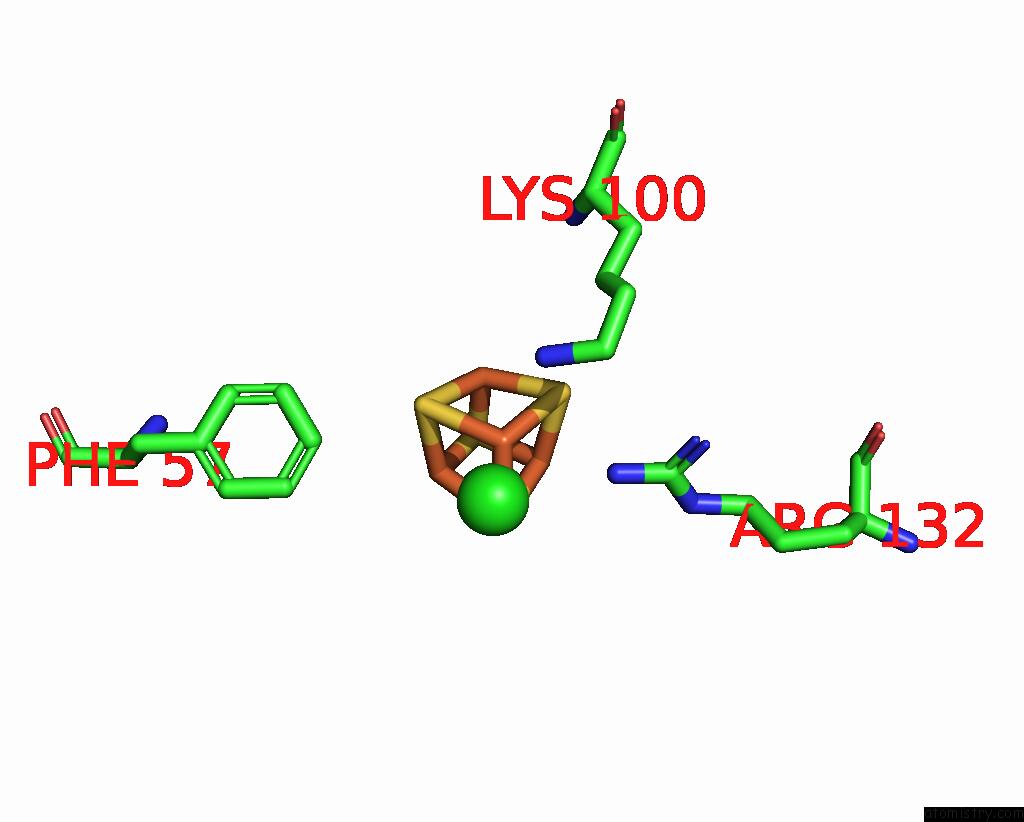
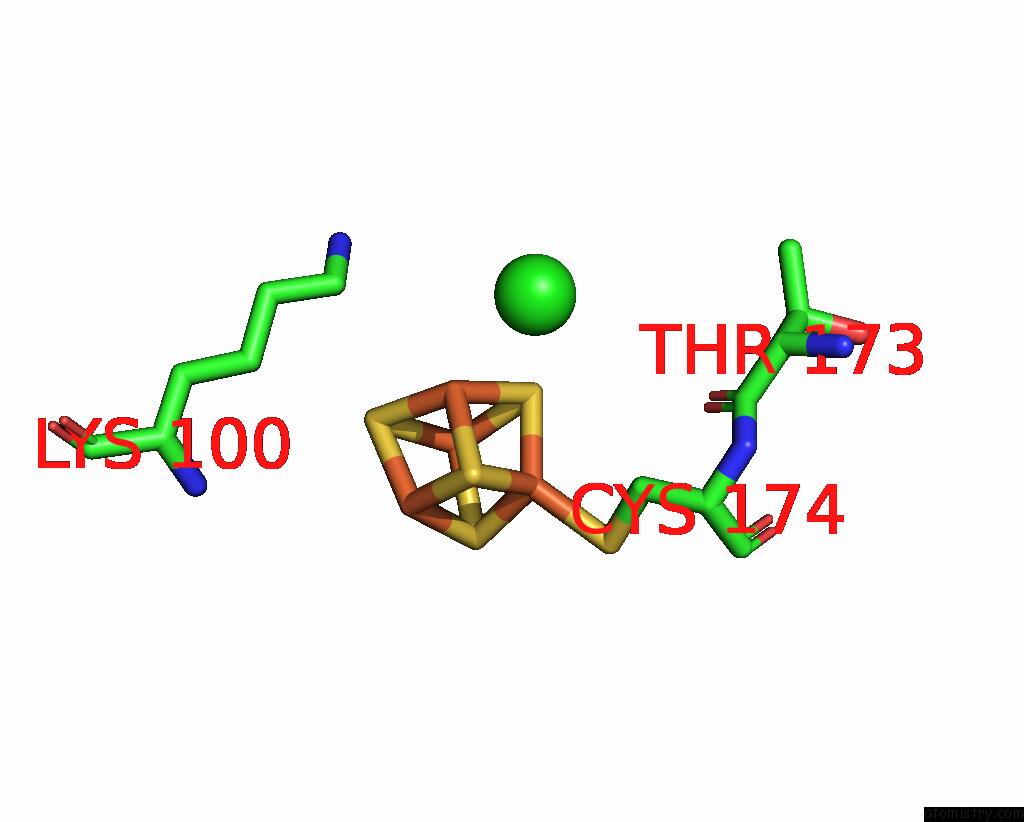
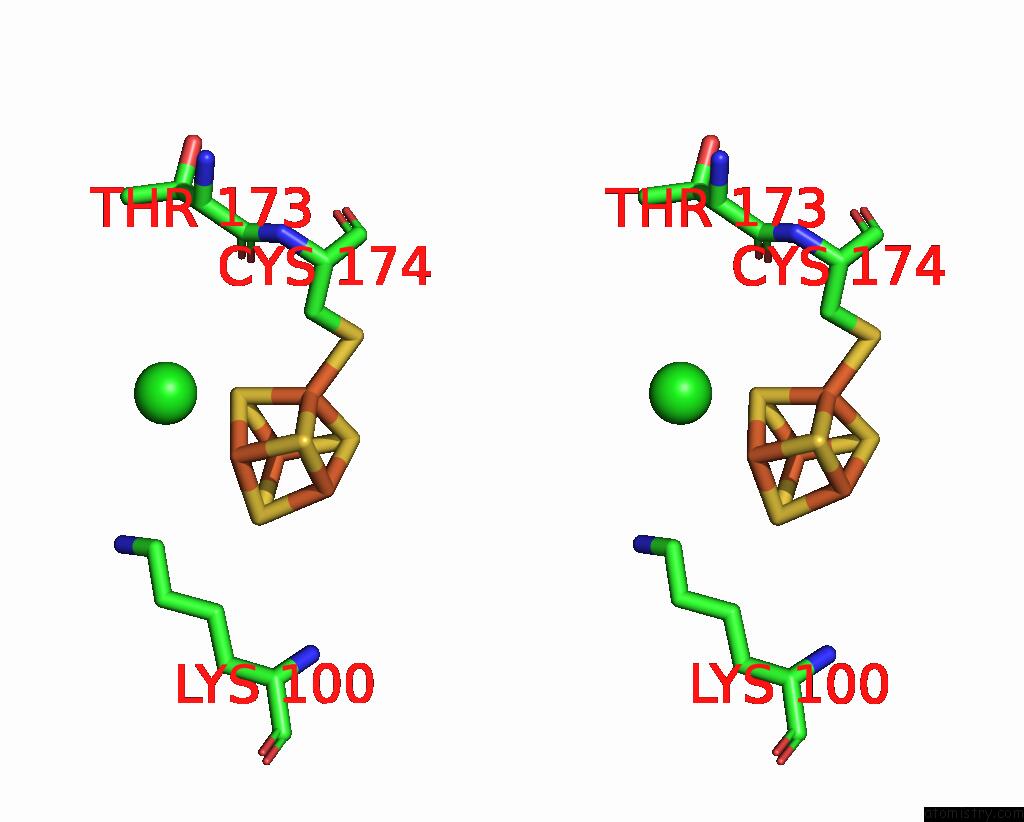
Chlorine binding site 3 out of 5 in 8cnz
Go back to
Chlorine binding site 3 out
of 5 in the Mmlare-[4FE-4S] Phased By Fe-Sad
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Mmlare-[4FE-4S] Phased By Fe-Sad within 5.0Å range:
|
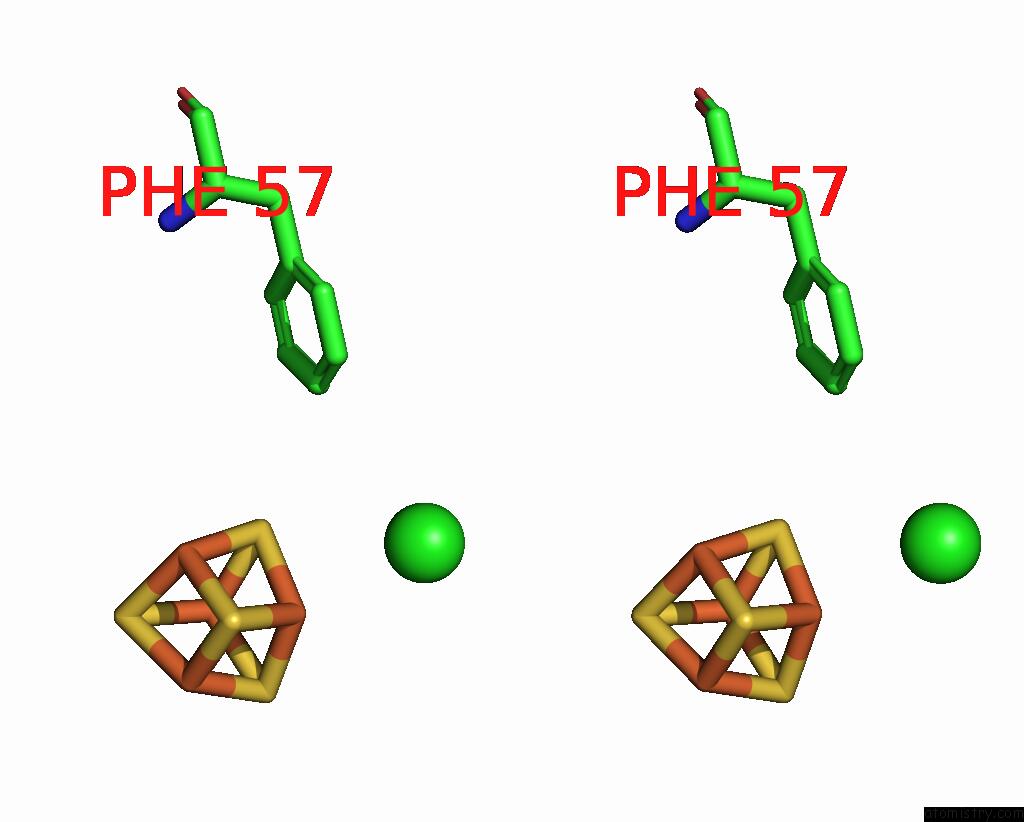
Chlorine binding site 4 out of 5 in 8cnz
Go back to
Chlorine binding site 4 out
of 5 in the Mmlare-[4FE-4S] Phased By Fe-Sad
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Mmlare-[4FE-4S] Phased By Fe-Sad within 5.0Å range:
|
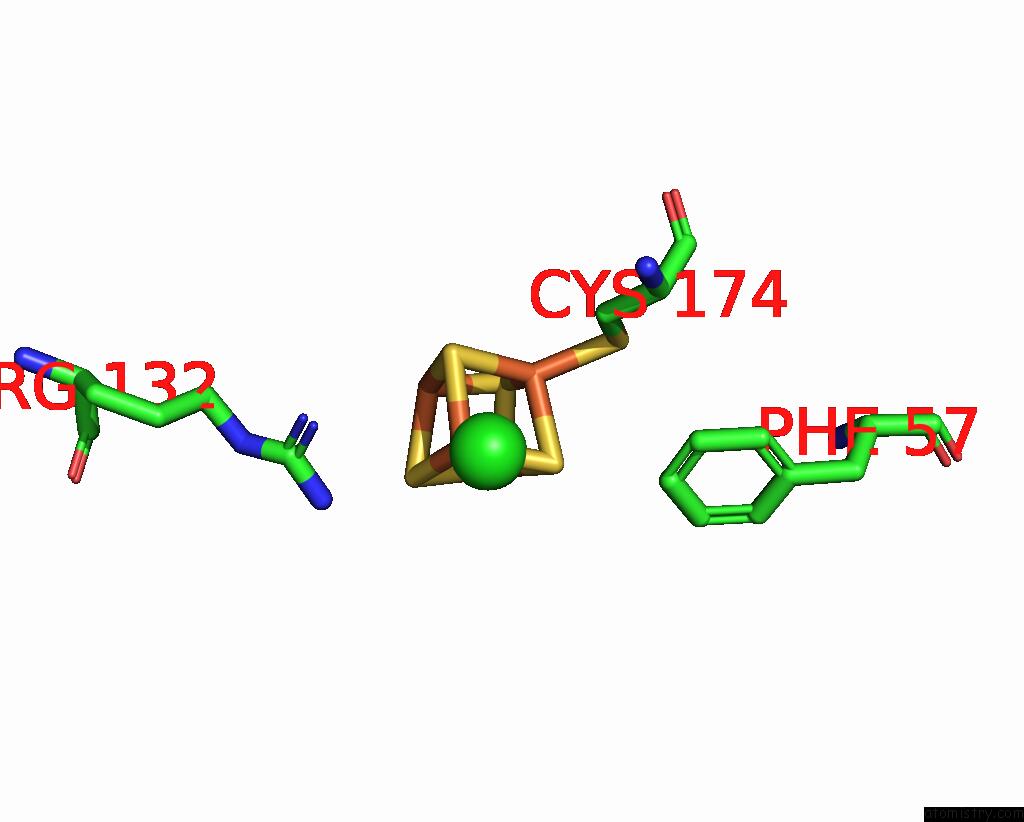
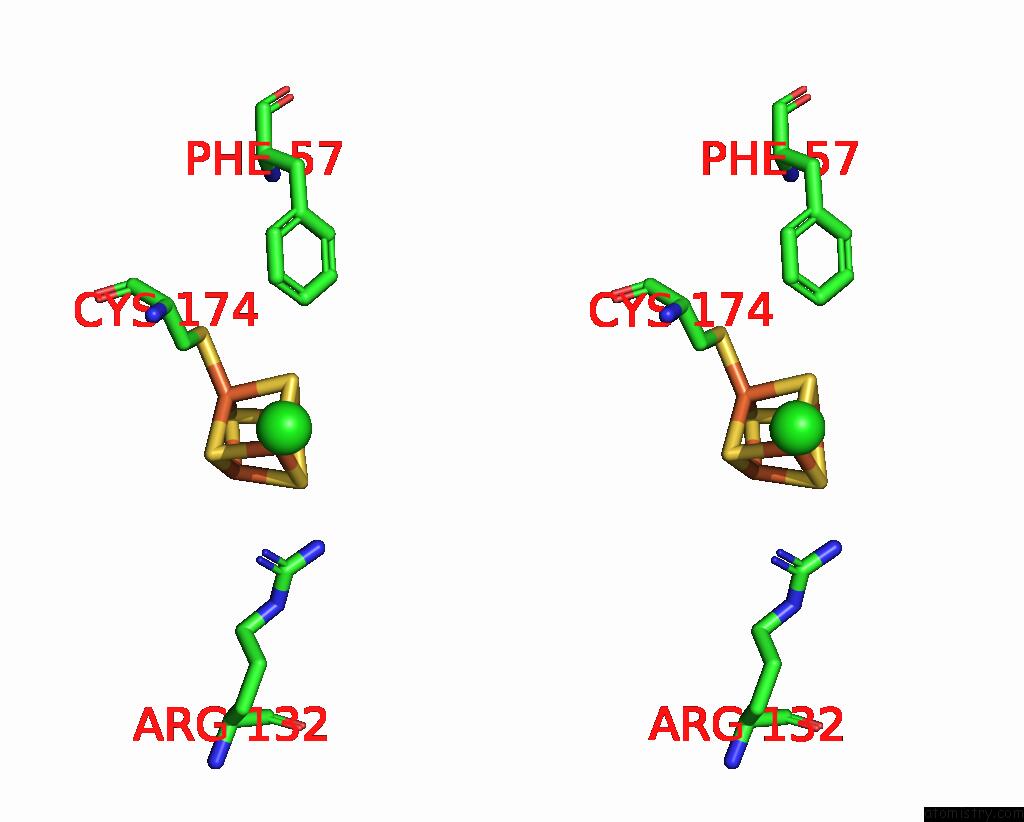
Chlorine binding site 5 out of 5 in 8cnz
Go back to
Chlorine binding site 5 out
of 5 in the Mmlare-[4FE-4S] Phased By Fe-Sad
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Mmlare-[4FE-4S] Phased By Fe-Sad within 5.0Å range:
|
Reference:
P.Zecchin,
L.Pecqueur,
J.Oltmanns,
C.Velours,
V.Schunemann,
M.Fontecave,
B.Golinelli-Pimpaneau.
Structure-Based Insights Into the Mechanism of [4FE-4S]-Dependent Sulfur Insertase Lare. Protein Sci. E4874 2023.
ISSN: ESSN 1469-896X
PubMed: 38100250
DOI: 10.1002/PRO.4874
Page generated: Sun Jul 13 10:15:51 2025
ISSN: ESSN 1469-896X
PubMed: 38100250
DOI: 10.1002/PRO.4874
Last articles
F in 7MONF in 7MOG
F in 7MOO
F in 7MML
F in 7MMI
F in 7MMK
F in 7MMJ
F in 7MMG
F in 7MMF
F in 7MMH