Chlorine »
PDB 8ckq-8ctm »
8ctb »
Chlorine in PDB 8ctb: Human PRMT5:MEP50 Structure with Fragment 3 and Mta Bound
Enzymatic activity of Human PRMT5:MEP50 Structure with Fragment 3 and Mta Bound
All present enzymatic activity of Human PRMT5:MEP50 Structure with Fragment 3 and Mta Bound:
2.1.1.320;
2.1.1.320;
Protein crystallography data
The structure of Human PRMT5:MEP50 Structure with Fragment 3 and Mta Bound, PDB code: 8ctb
was solved by
R.J.Gunn,
J.D.Lawson,
C.R.Smith,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 44.67 / 2.61 |
| Space group | I 2 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 97.983, 137.232, 178.304, 90, 90, 90 |
| R / Rfree (%) | 24.7 / 28.9 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Human PRMT5:MEP50 Structure with Fragment 3 and Mta Bound
(pdb code 8ctb). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Human PRMT5:MEP50 Structure with Fragment 3 and Mta Bound, PDB code: 8ctb:
In total only one binding site of Chlorine was determined in the Human PRMT5:MEP50 Structure with Fragment 3 and Mta Bound, PDB code: 8ctb:
Chlorine binding site 1 out of 1 in 8ctb
Go back to
Chlorine binding site 1 out
of 1 in the Human PRMT5:MEP50 Structure with Fragment 3 and Mta Bound
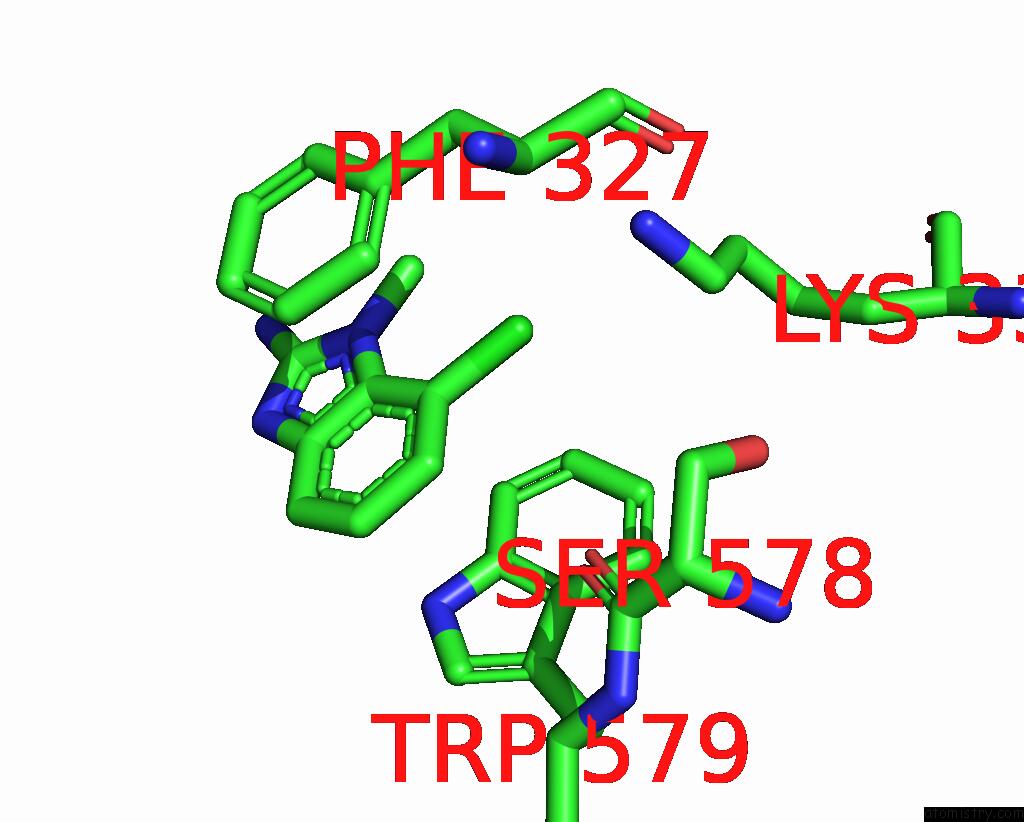
Mono view
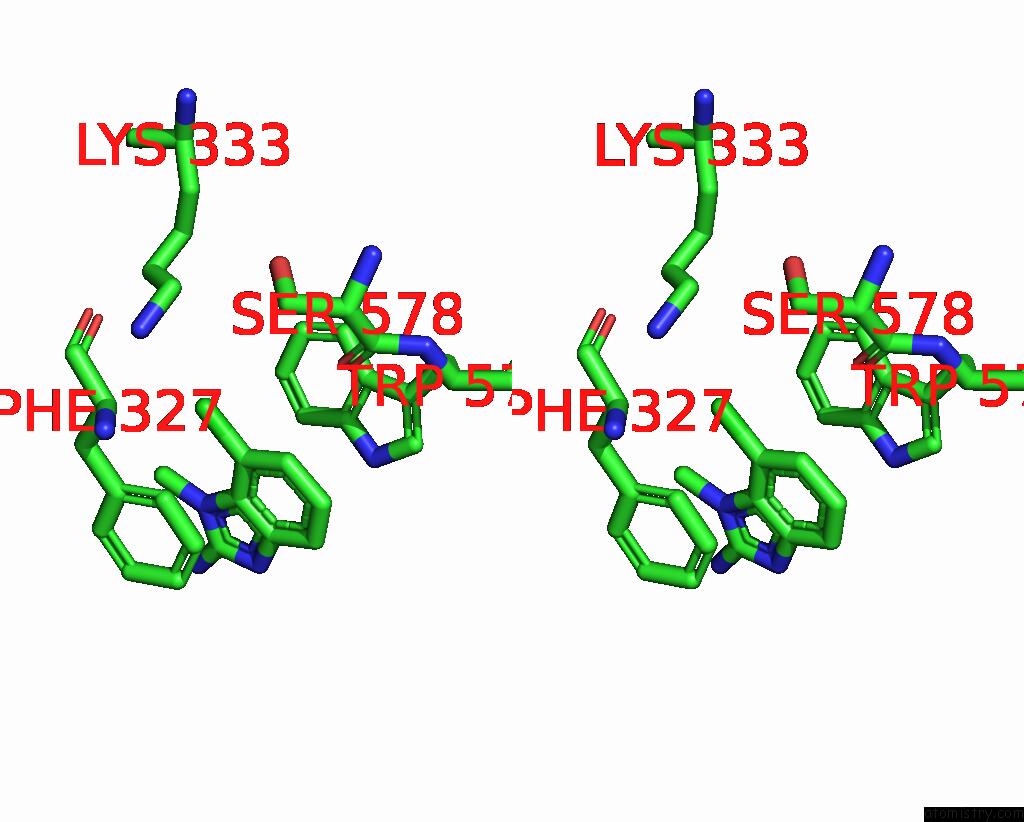
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Human PRMT5:MEP50 Structure with Fragment 3 and Mta Bound within 5.0Å range:
|
Reference:
C.R.Smith,
S.Kulyk,
M.U.D.Ahmad,
V.Arkhipova,
J.G.Christensen,
R.J.Gunn,
A.Ivetac,
J.M.Ketcham,
J.Kuehler,
J.D.Lawson,
N.C.Thomas,
X.Wang,
M.A.Marx.
Fragment Optimization and Elaboration Strategies - the Discovery of Two Lead Series of PRMT5/Mta Inhibitors From Five Fragment Hits Rsc Med Chem 2022.
ISSN: ESSN 2632-8682
DOI: 10.1039/D2MD00163B
Page generated: Sun Jul 13 10:20:21 2025
ISSN: ESSN 2632-8682
DOI: 10.1039/D2MD00163B
Last articles
Co in 1M77Co in 1M5A
Co in 1JYM
Co in 1LNA
Co in 1LFM
Co in 1L8X
Co in 1KEJ
Co in 1KC2
Co in 1K98
Co in 1K7Y