Chlorine »
PDB 8f4y-8fh7 »
8fc8 »
Chlorine in PDB 8fc8: Cryo-Em Structure of the Human TRPV4 in Complex with GSK1016790A
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Cryo-Em Structure of the Human TRPV4 in Complex with GSK1016790A
(pdb code 8fc8). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 8 binding sites of Chlorine where determined in the Cryo-Em Structure of the Human TRPV4 in Complex with GSK1016790A, PDB code: 8fc8:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Chlorine where determined in the Cryo-Em Structure of the Human TRPV4 in Complex with GSK1016790A, PDB code: 8fc8:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Chlorine binding site 1 out of 8 in 8fc8
Go back to
Chlorine binding site 1 out
of 8 in the Cryo-Em Structure of the Human TRPV4 in Complex with GSK1016790A

Mono view
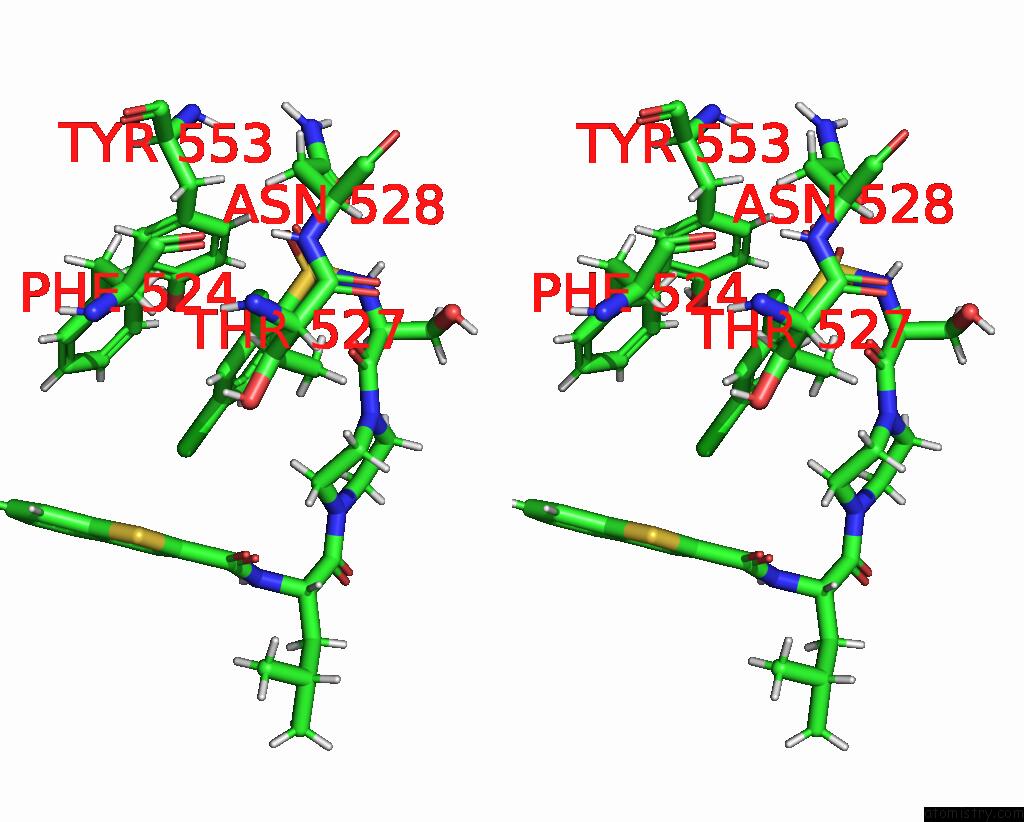
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Cryo-Em Structure of the Human TRPV4 in Complex with GSK1016790A within 5.0Å range:
|
Chlorine binding site 2 out of 8 in 8fc8
Go back to
Chlorine binding site 2 out
of 8 in the Cryo-Em Structure of the Human TRPV4 in Complex with GSK1016790A

Mono view
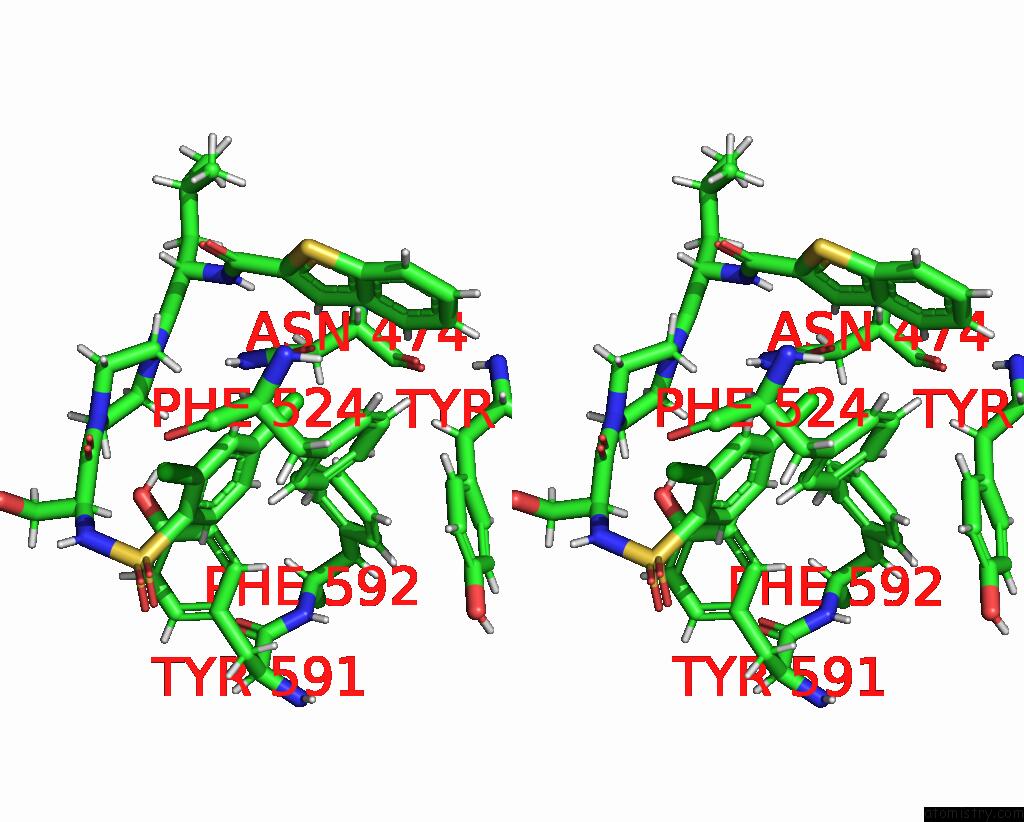
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Cryo-Em Structure of the Human TRPV4 in Complex with GSK1016790A within 5.0Å range:
|
Chlorine binding site 3 out of 8 in 8fc8
Go back to
Chlorine binding site 3 out
of 8 in the Cryo-Em Structure of the Human TRPV4 in Complex with GSK1016790A
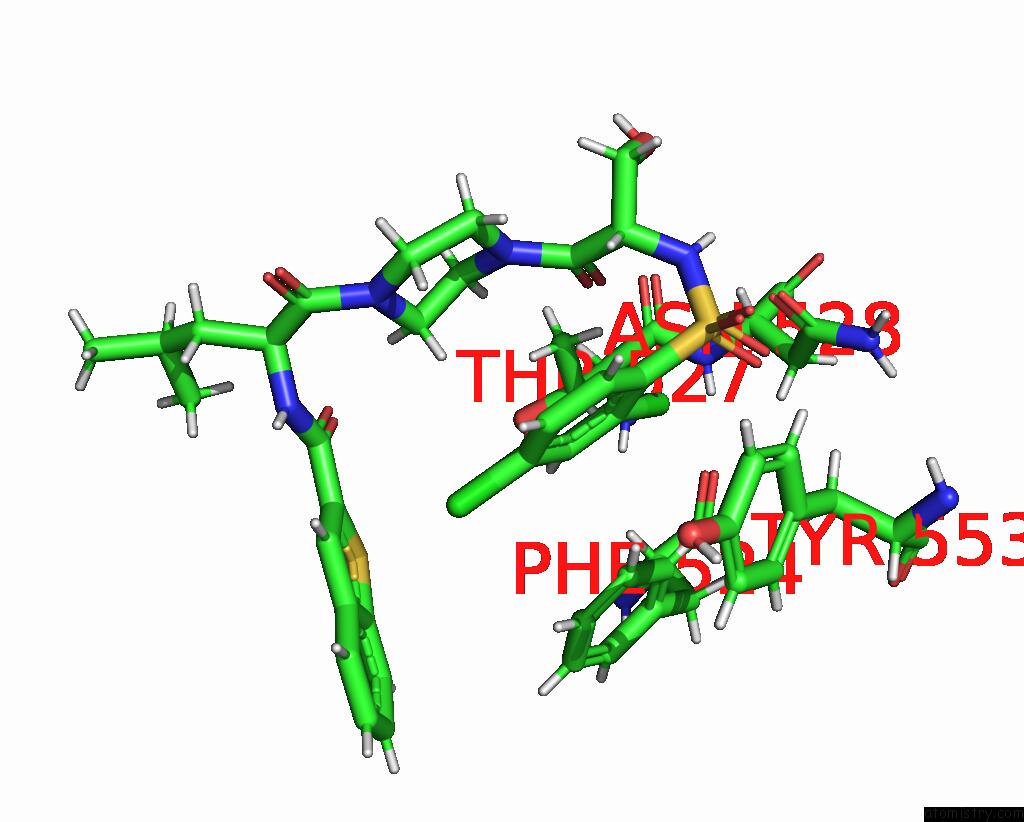
Mono view
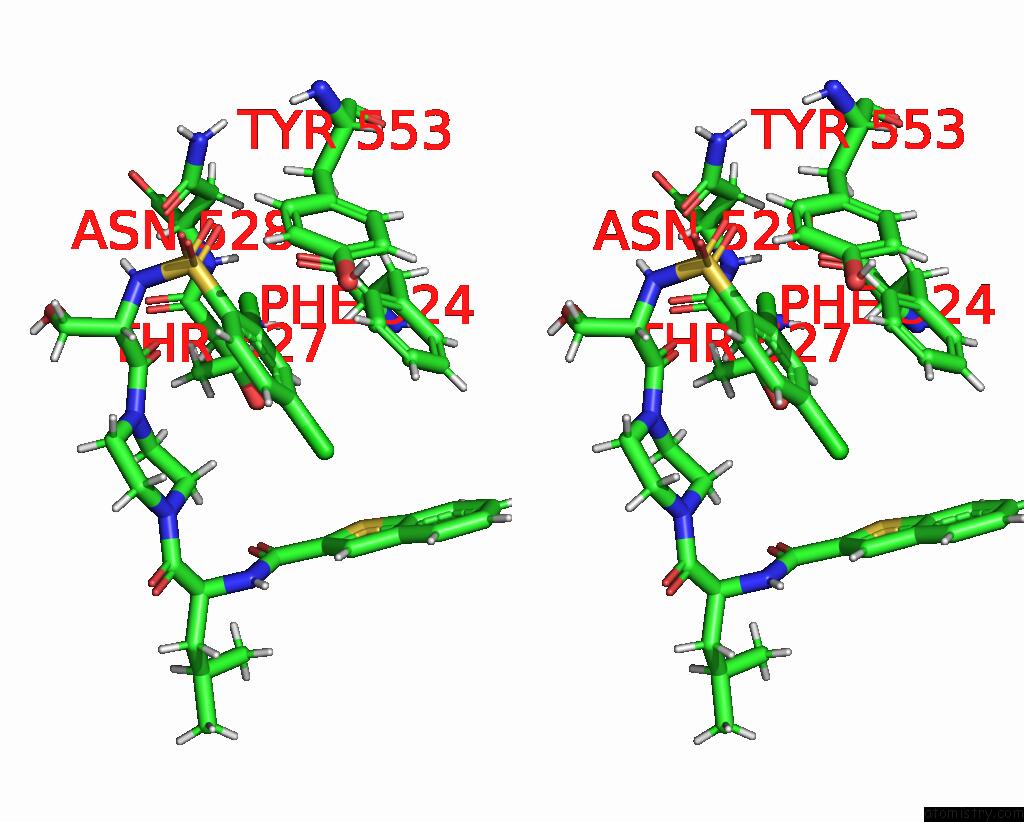
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Cryo-Em Structure of the Human TRPV4 in Complex with GSK1016790A within 5.0Å range:
|
Chlorine binding site 4 out of 8 in 8fc8
Go back to
Chlorine binding site 4 out
of 8 in the Cryo-Em Structure of the Human TRPV4 in Complex with GSK1016790A

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Cryo-Em Structure of the Human TRPV4 in Complex with GSK1016790A within 5.0Å range:
|
Chlorine binding site 5 out of 8 in 8fc8
Go back to
Chlorine binding site 5 out
of 8 in the Cryo-Em Structure of the Human TRPV4 in Complex with GSK1016790A
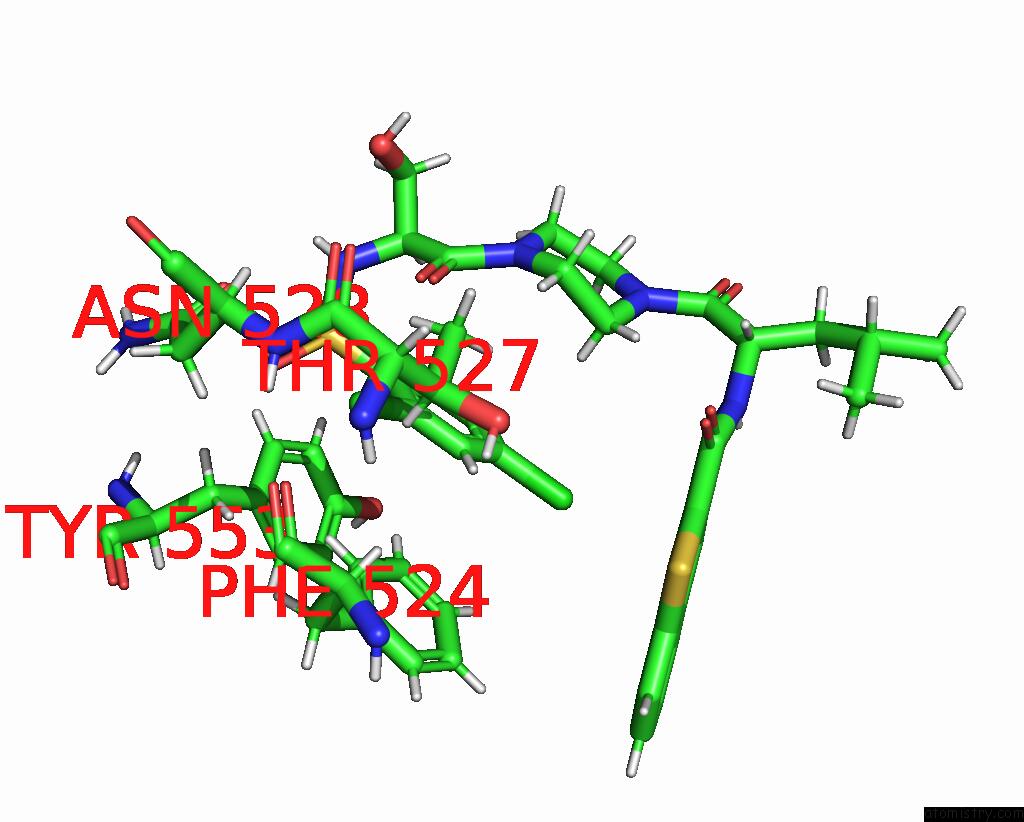
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Cryo-Em Structure of the Human TRPV4 in Complex with GSK1016790A within 5.0Å range:
|
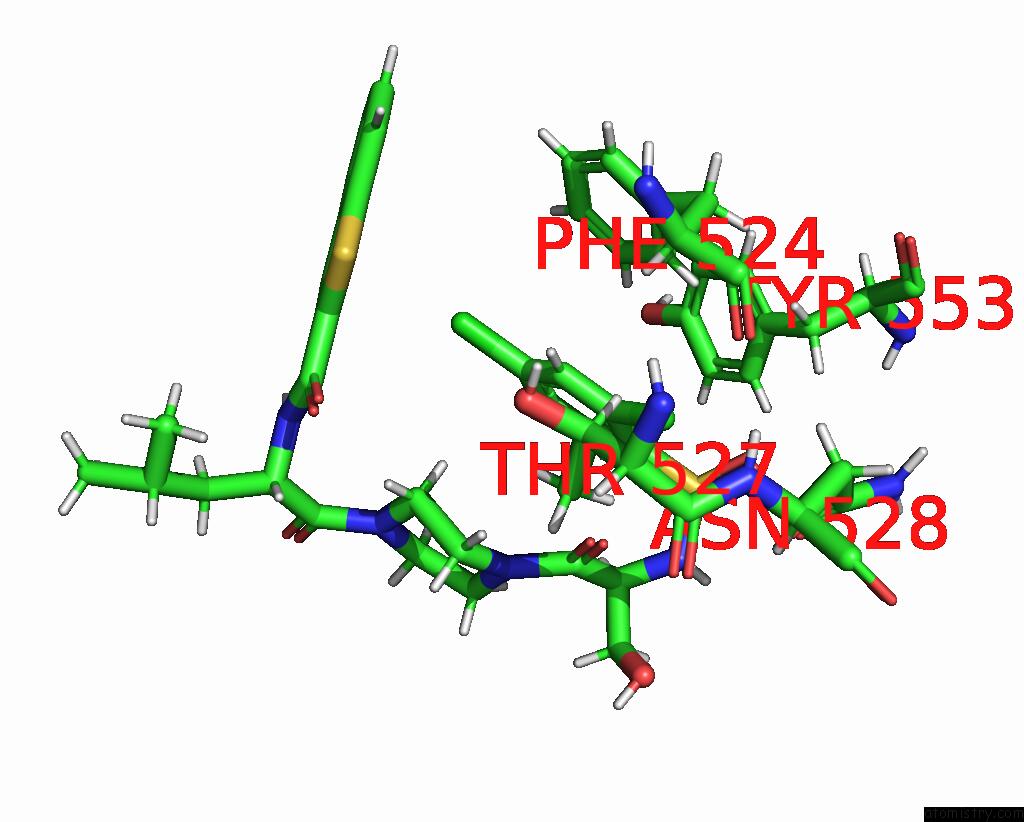
Chlorine binding site 6 out of 8 in 8fc8
Go back to
Chlorine binding site 6 out
of 8 in the Cryo-Em Structure of the Human TRPV4 in Complex with GSK1016790A
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Cryo-Em Structure of the Human TRPV4 in Complex with GSK1016790A within 5.0Å range:
|
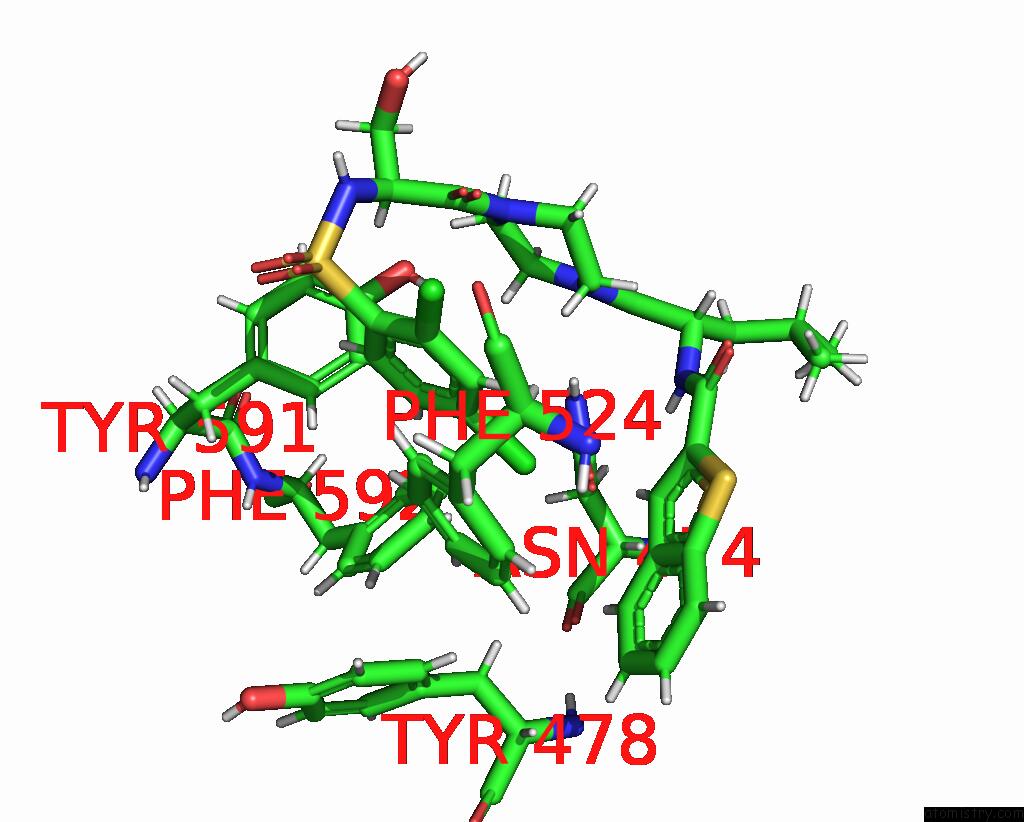
Chlorine binding site 7 out of 8 in 8fc8
Go back to
Chlorine binding site 7 out
of 8 in the Cryo-Em Structure of the Human TRPV4 in Complex with GSK1016790A
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 7 of Cryo-Em Structure of the Human TRPV4 in Complex with GSK1016790A within 5.0Å range:
|
Chlorine binding site 8 out of 8 in 8fc8
Go back to
Chlorine binding site 8 out
of 8 in the Cryo-Em Structure of the Human TRPV4 in Complex with GSK1016790A

Mono view
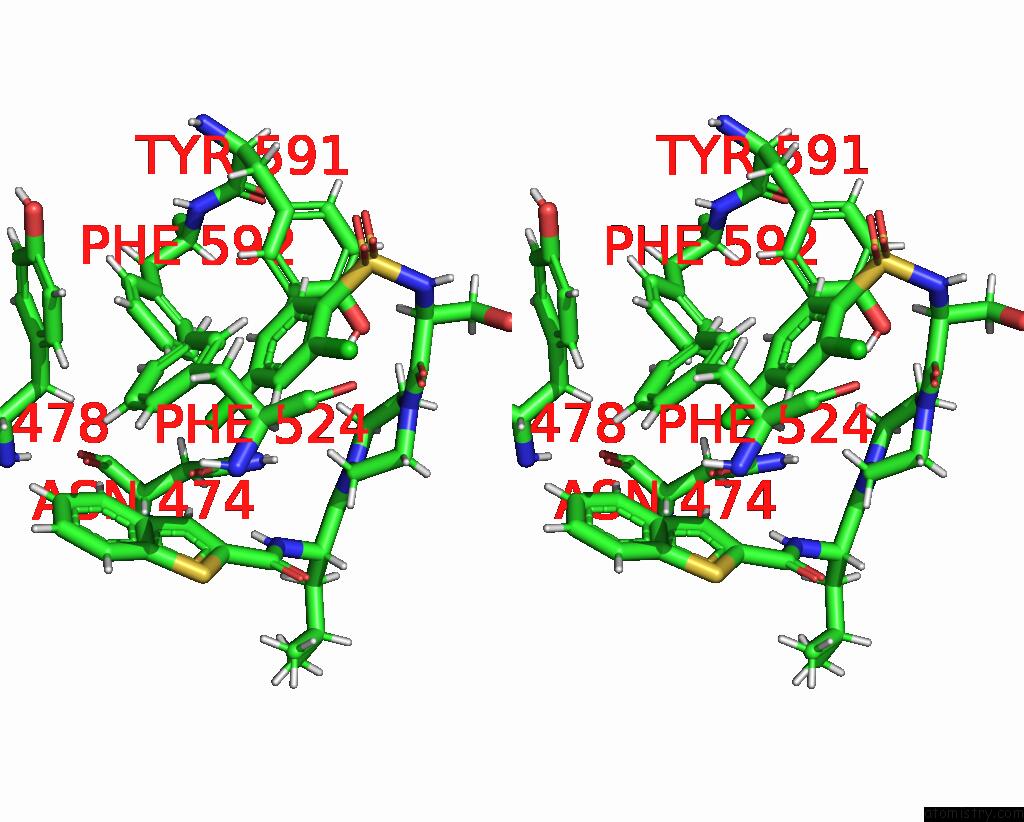
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 8 of Cryo-Em Structure of the Human TRPV4 in Complex with GSK1016790A within 5.0Å range:
|
Reference:
D.H.Kwon,
F.Zhang,
B.A.Mccray,
S.Feng,
M.Kumar,
J.M.Sullivan,
W.Im,
C.J.Sumner,
S.Y.Lee.
TRPV4-Rho Gtpase Complex Structures Reveal Mechanisms of Gating and Disease. Nat Commun V. 14 3732 2023.
ISSN: ESSN 2041-1723
PubMed: 37353484
DOI: 10.1038/S41467-023-39345-0
Page generated: Sun Jul 13 11:21:07 2025
ISSN: ESSN 2041-1723
PubMed: 37353484
DOI: 10.1038/S41467-023-39345-0
Last articles
Fe in 2YXOFe in 2YRS
Fe in 2YXC
Fe in 2YNM
Fe in 2YVJ
Fe in 2YP1
Fe in 2YU2
Fe in 2YU1
Fe in 2YQB
Fe in 2YOO