Chlorine »
PDB 8fh8-8fpl »
8flg »
Chlorine in PDB 8flg: Bruton'S Tyrosine Kinase in Complex with An Orthosteric Inhibitor
Enzymatic activity of Bruton'S Tyrosine Kinase in Complex with An Orthosteric Inhibitor
All present enzymatic activity of Bruton'S Tyrosine Kinase in Complex with An Orthosteric Inhibitor:
2.7.10.2;
2.7.10.2;
Protein crystallography data
The structure of Bruton'S Tyrosine Kinase in Complex with An Orthosteric Inhibitor, PDB code: 8flg
was solved by
C.M.Metrick,
D.J.Marcotte,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 31.23 / 2.20 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 71.241, 104.24, 38.018, 90, 90, 90 |
| R / Rfree (%) | 19.5 / 25.4 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Bruton'S Tyrosine Kinase in Complex with An Orthosteric Inhibitor
(pdb code 8flg). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Bruton'S Tyrosine Kinase in Complex with An Orthosteric Inhibitor, PDB code: 8flg:
In total only one binding site of Chlorine was determined in the Bruton'S Tyrosine Kinase in Complex with An Orthosteric Inhibitor, PDB code: 8flg:
Chlorine binding site 1 out of 1 in 8flg
Go back to
Chlorine binding site 1 out
of 1 in the Bruton'S Tyrosine Kinase in Complex with An Orthosteric Inhibitor

Mono view
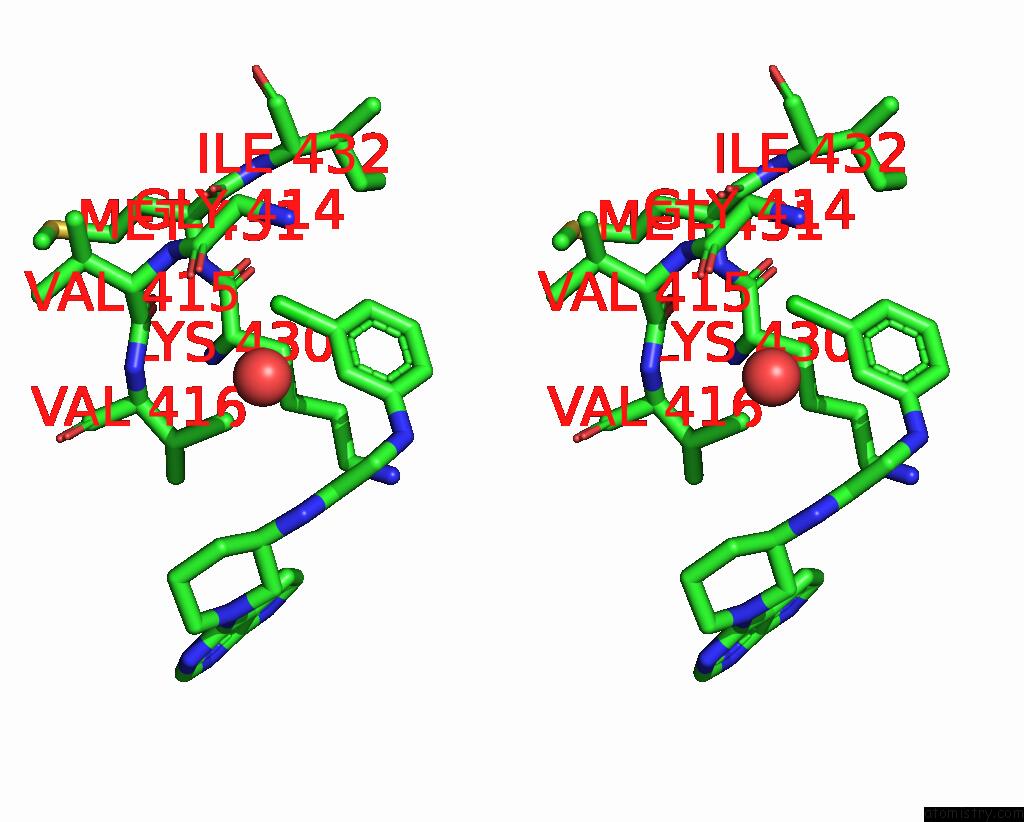
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Bruton'S Tyrosine Kinase in Complex with An Orthosteric Inhibitor within 5.0Å range:
|
Reference:
G.H.Vandeveer,
R.M.Arduini,
D.P.Baker,
K.Barry,
T.Bohnert,
J.K.Bowden-Verhoek,
P.Conlon,
P.F.Cullen,
B.Guan,
T.J.Jenkins,
S.Y.Liao,
L.Lin,
Y.T.Liu,
D.Marcotte,
E.Mertsching,
C.M.Metrick,
E.Negrou,
N.Powell,
D.Scott,
L.F.Silvian,
B.T.Hopkins.
Discovery of Structural Diverse Reversible Btk Inhibitors Utilized to Develop A Novel in Vivo CD69 and CD86 Pk/Pd Mouse Model. Bioorg.Med.Chem.Lett. V. 80 29108 2023.
ISSN: ESSN 1464-3405
PubMed: 36538993
DOI: 10.1016/J.BMCL.2022.129108
Page generated: Sun Jul 13 11:28:44 2025
ISSN: ESSN 1464-3405
PubMed: 36538993
DOI: 10.1016/J.BMCL.2022.129108
Last articles
Co in 2G6FCo in 2FQN
Co in 2FUA
Co in 2FQT
Co in 2FIJ
Co in 2FQO
Co in 2FGP
Co in 2EVO
Co in 2F7N
Co in 2FDF