Chlorine in PDB 8gw7: ATSLAC1 6D Mutant in Open State
Chlorine Binding Sites:
The binding sites of Chlorine atom in the ATSLAC1 6D Mutant in Open State
(pdb code 8gw7). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 6 binding sites of Chlorine where determined in the ATSLAC1 6D Mutant in Open State, PDB code: 8gw7:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Chlorine where determined in the ATSLAC1 6D Mutant in Open State, PDB code: 8gw7:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
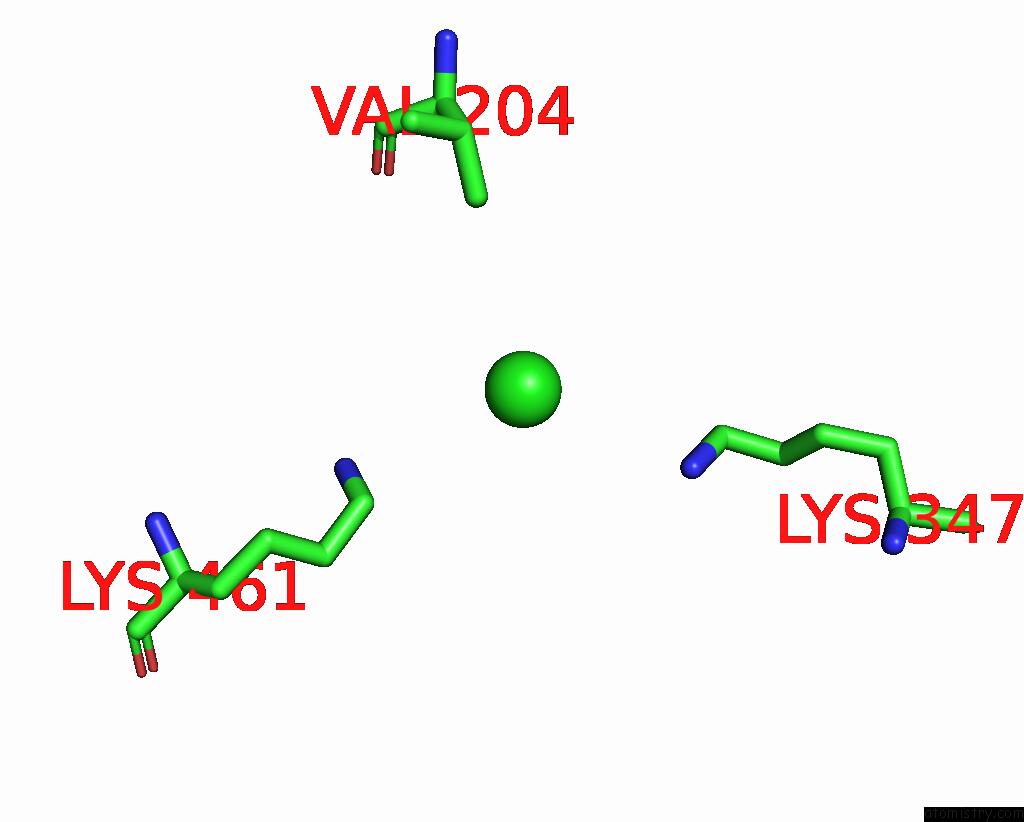
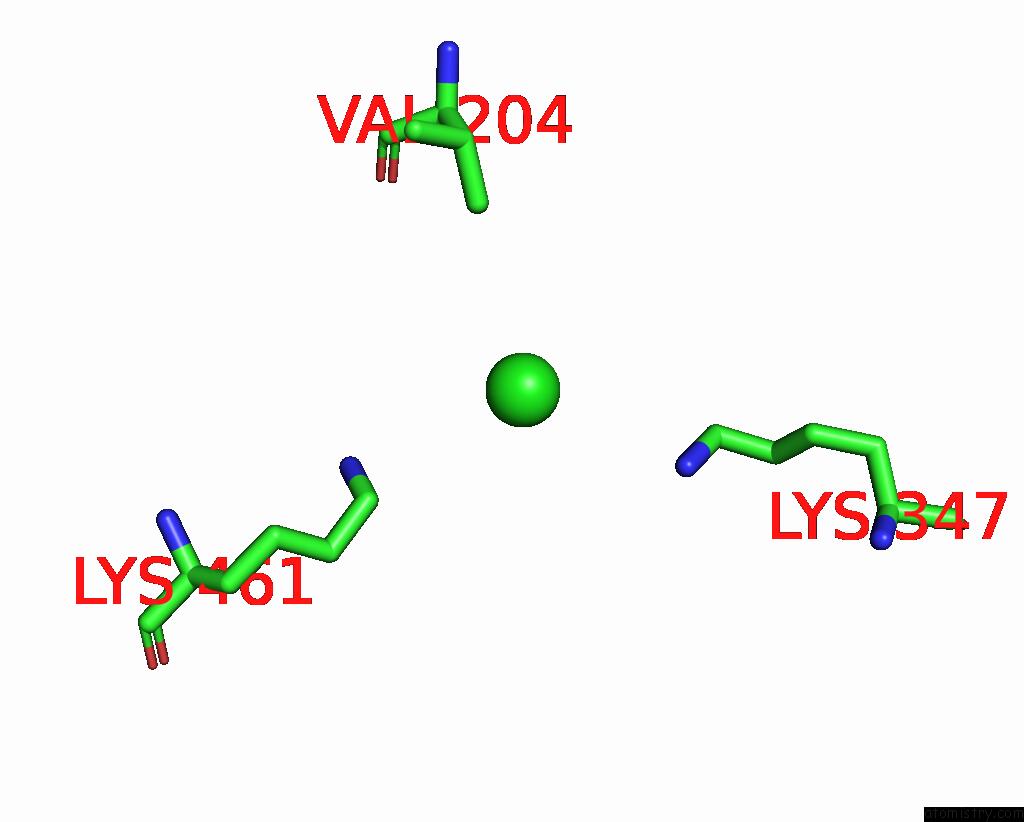
Chlorine binding site 1 out of 6 in 8gw7
Go back to
Chlorine binding site 1 out
of 6 in the ATSLAC1 6D Mutant in Open State
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of ATSLAC1 6D Mutant in Open State within 5.0Å range:
|
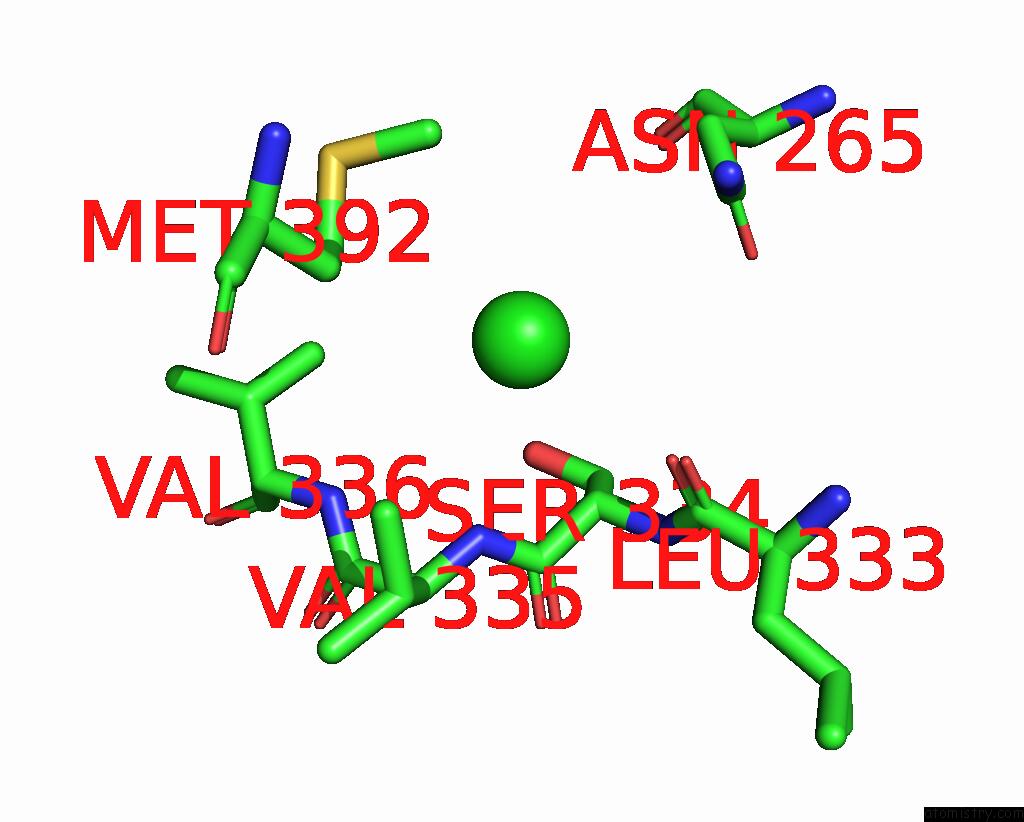
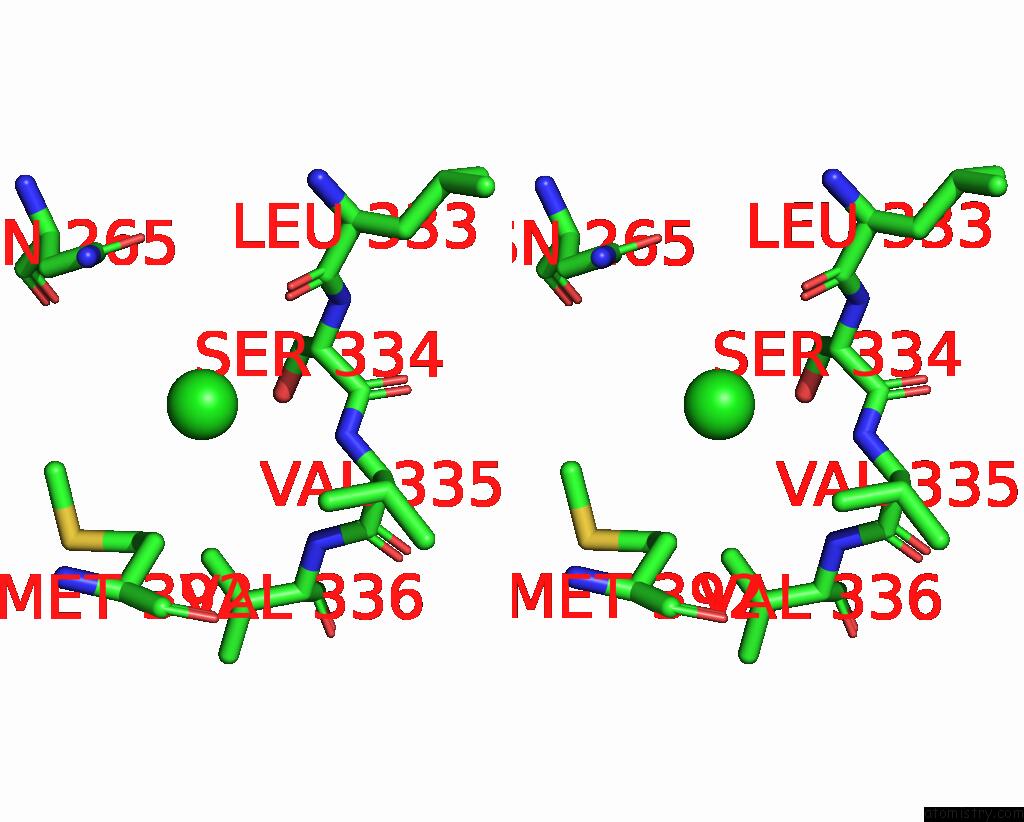
Chlorine binding site 2 out of 6 in 8gw7
Go back to
Chlorine binding site 2 out
of 6 in the ATSLAC1 6D Mutant in Open State
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of ATSLAC1 6D Mutant in Open State within 5.0Å range:
|
Chlorine binding site 3 out of 6 in 8gw7
Go back to
Chlorine binding site 3 out
of 6 in the ATSLAC1 6D Mutant in Open State
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of ATSLAC1 6D Mutant in Open State within 5.0Å range:
|
Chlorine binding site 4 out of 6 in 8gw7
Go back to
Chlorine binding site 4 out
of 6 in the ATSLAC1 6D Mutant in Open State

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of ATSLAC1 6D Mutant in Open State within 5.0Å range:
|
Chlorine binding site 5 out of 6 in 8gw7
Go back to
Chlorine binding site 5 out
of 6 in the ATSLAC1 6D Mutant in Open State
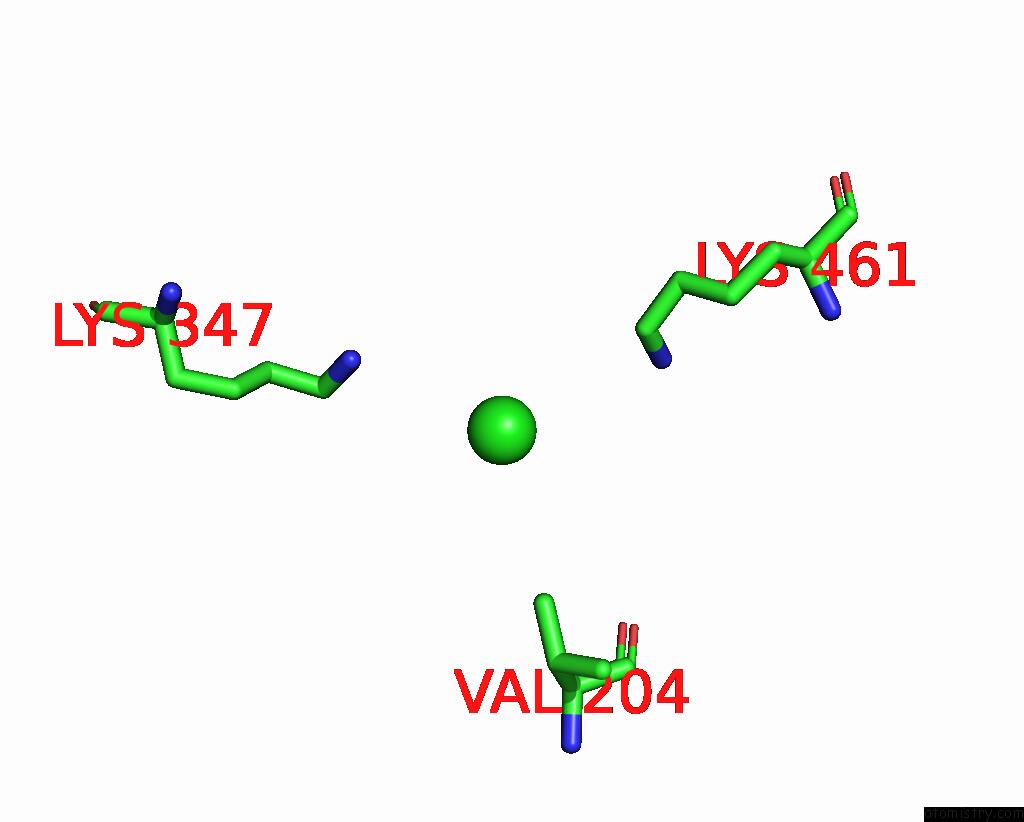
Mono view
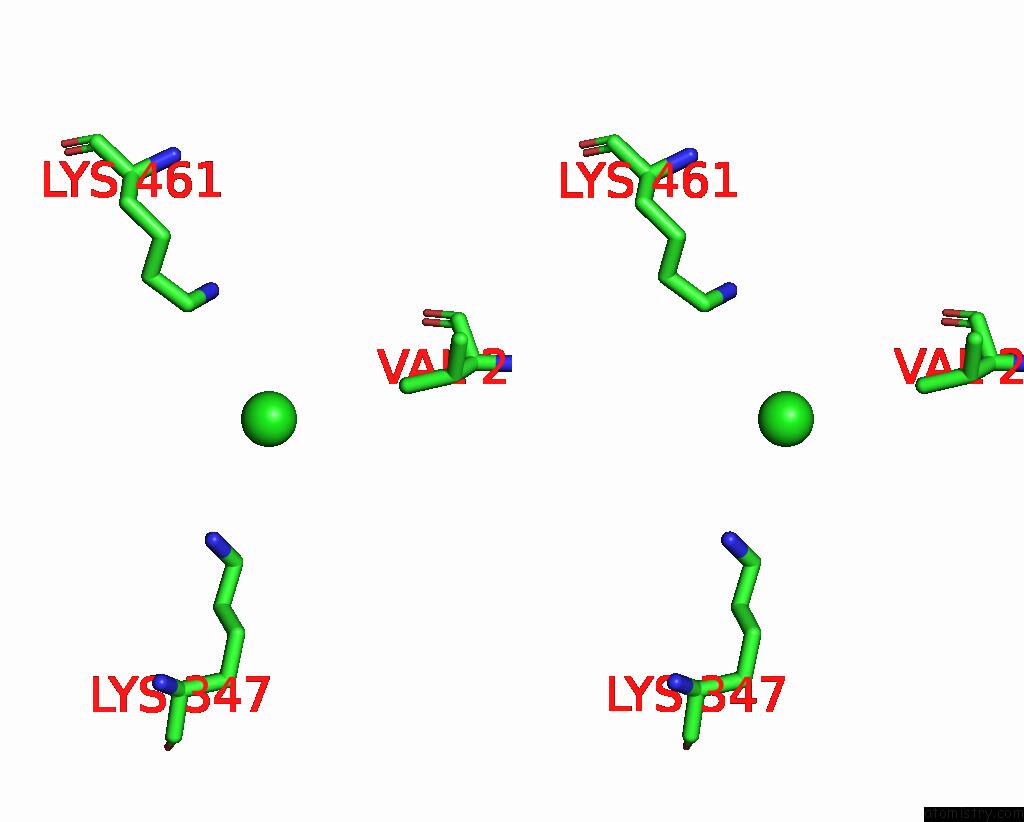
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of ATSLAC1 6D Mutant in Open State within 5.0Å range:
|
Chlorine binding site 6 out of 6 in 8gw7
Go back to
Chlorine binding site 6 out
of 6 in the ATSLAC1 6D Mutant in Open State
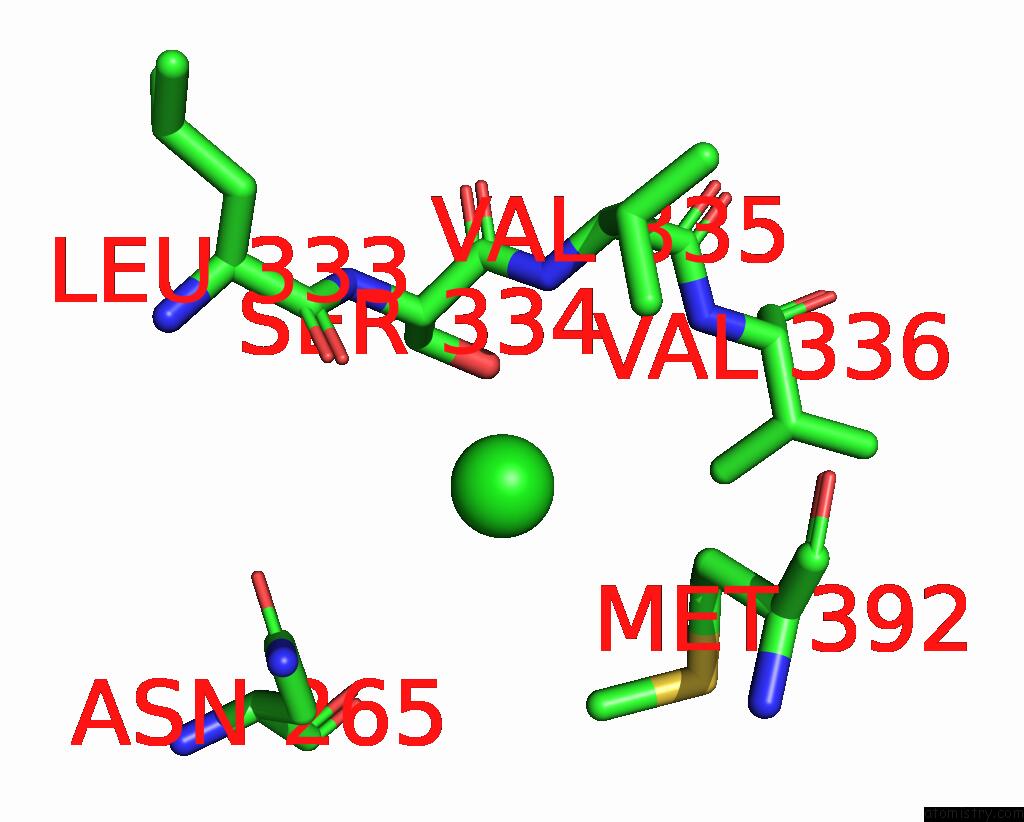
Mono view
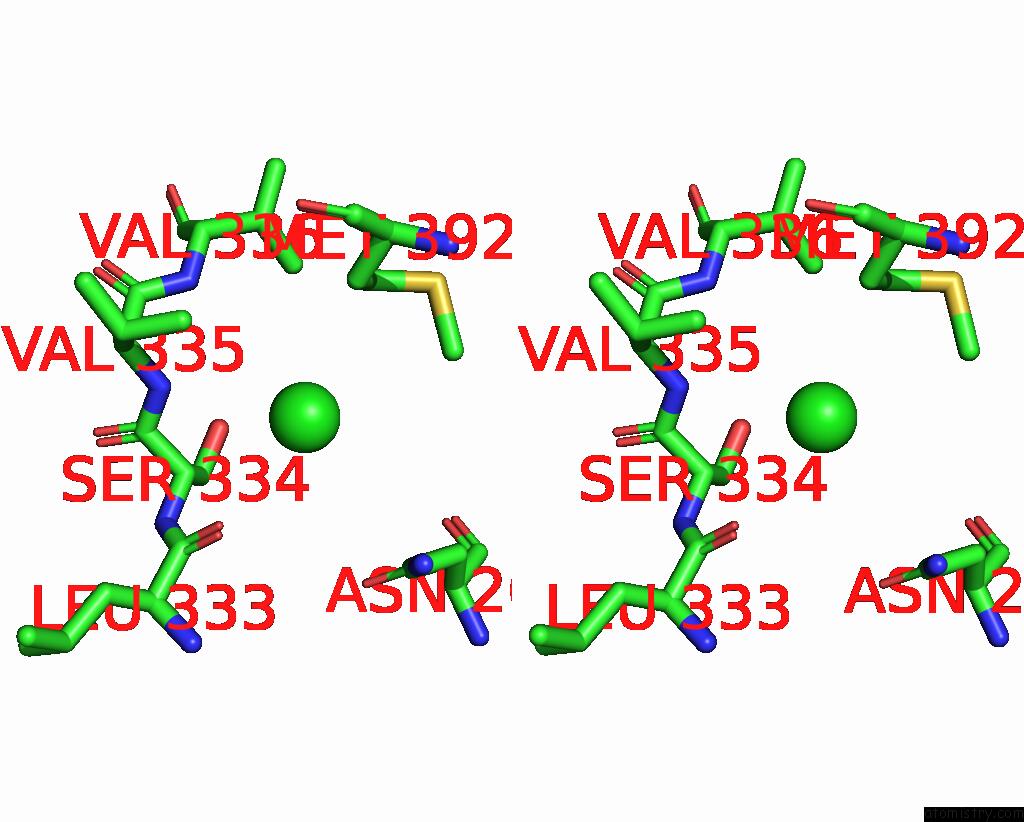
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of ATSLAC1 6D Mutant in Open State within 5.0Å range:
|
Reference:
Y.Lee,
S.Lee.
Cryo-Em Structures of the Plant Anion Channel SLAC1 From Arabidopsis Thaliana Suggest A Combined Activation Model To Be Published.
DOI: 10.1038/S41467-023-43193-3
Page generated: Tue Jul 30 10:10:12 2024
DOI: 10.1038/S41467-023-43193-3
Last articles
Zn in 9JPJZn in 9JP7
Zn in 9JPK
Zn in 9JPL
Zn in 9GN6
Zn in 9GN7
Zn in 9GKU
Zn in 9GKW
Zn in 9GKX
Zn in 9GL0