Chlorine »
PDB 8pm6-8pz0 »
8poz »
Chlorine in PDB 8poz: Crystal Structure of the C120G Variant of the Membrane-Bound [Nife]- Hydrogenase From Cupriavidus Necator in the H2-Reduced State at 1.65 A Resolution.
Enzymatic activity of Crystal Structure of the C120G Variant of the Membrane-Bound [Nife]- Hydrogenase From Cupriavidus Necator in the H2-Reduced State at 1.65 A Resolution.
All present enzymatic activity of Crystal Structure of the C120G Variant of the Membrane-Bound [Nife]- Hydrogenase From Cupriavidus Necator in the H2-Reduced State at 1.65 A Resolution.:
1.12.99.6;
1.12.99.6;
Protein crystallography data
The structure of Crystal Structure of the C120G Variant of the Membrane-Bound [Nife]- Hydrogenase From Cupriavidus Necator in the H2-Reduced State at 1.65 A Resolution., PDB code: 8poz
was solved by
A.Schmidt,
J.Kalms,
P.Scheerer,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.92 / 1.65 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 73.415, 95.751, 120.597, 90, 90, 90 |
| R / Rfree (%) | 14.1 / 17.2 |
Other elements in 8poz:
The structure of Crystal Structure of the C120G Variant of the Membrane-Bound [Nife]- Hydrogenase From Cupriavidus Necator in the H2-Reduced State at 1.65 A Resolution. also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
| Iron | (Fe) | 16 atoms |
| Nickel | (Ni) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of the C120G Variant of the Membrane-Bound [Nife]- Hydrogenase From Cupriavidus Necator in the H2-Reduced State at 1.65 A Resolution.
(pdb code 8poz). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 4 binding sites of Chlorine where determined in the Crystal Structure of the C120G Variant of the Membrane-Bound [Nife]- Hydrogenase From Cupriavidus Necator in the H2-Reduced State at 1.65 A Resolution., PDB code: 8poz:
Jump to Chlorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Chlorine where determined in the Crystal Structure of the C120G Variant of the Membrane-Bound [Nife]- Hydrogenase From Cupriavidus Necator in the H2-Reduced State at 1.65 A Resolution., PDB code: 8poz:
Jump to Chlorine binding site number: 1; 2; 3; 4;
Chlorine binding site 1 out of 4 in 8poz
Go back to
Chlorine binding site 1 out
of 4 in the Crystal Structure of the C120G Variant of the Membrane-Bound [Nife]- Hydrogenase From Cupriavidus Necator in the H2-Reduced State at 1.65 A Resolution.
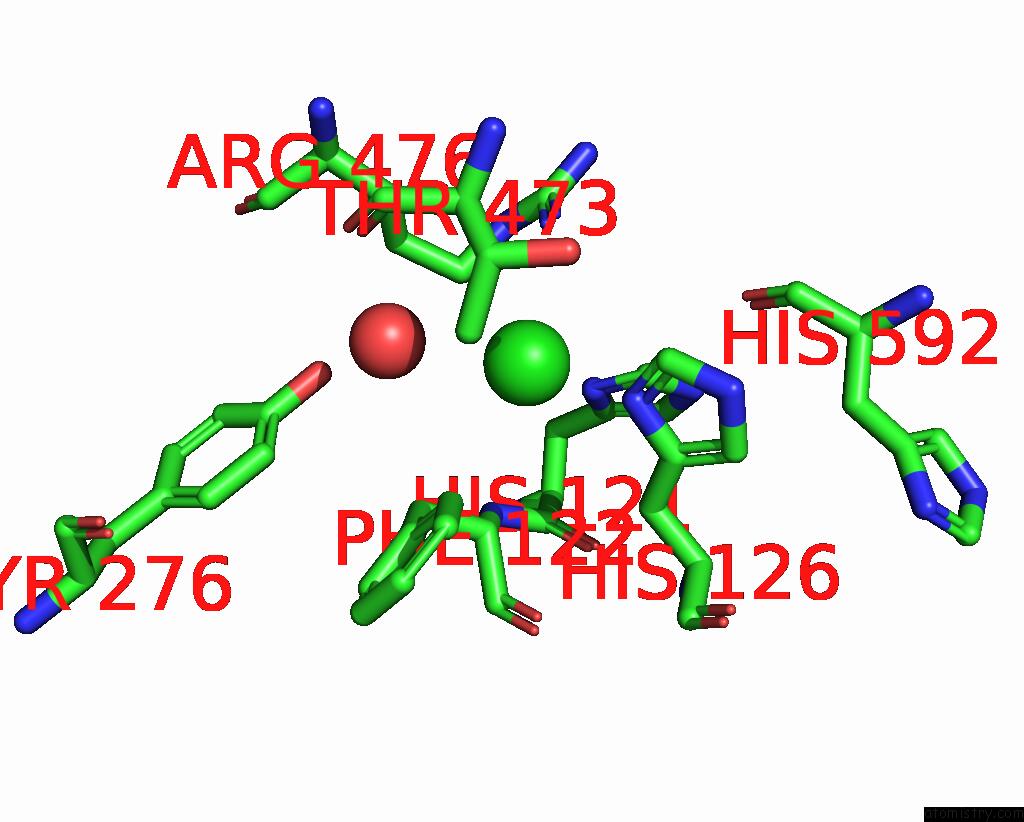
Mono view
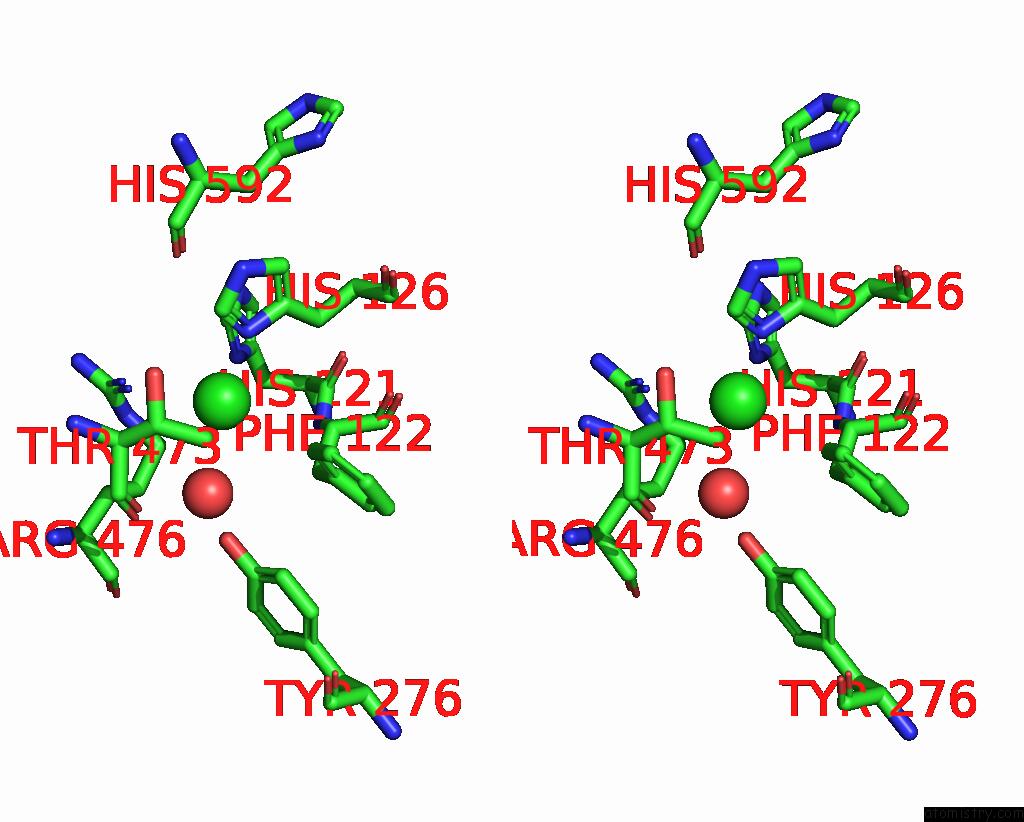
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of the C120G Variant of the Membrane-Bound [Nife]- Hydrogenase From Cupriavidus Necator in the H2-Reduced State at 1.65 A Resolution. within 5.0Å range:
|
Chlorine binding site 2 out of 4 in 8poz
Go back to
Chlorine binding site 2 out
of 4 in the Crystal Structure of the C120G Variant of the Membrane-Bound [Nife]- Hydrogenase From Cupriavidus Necator in the H2-Reduced State at 1.65 A Resolution.
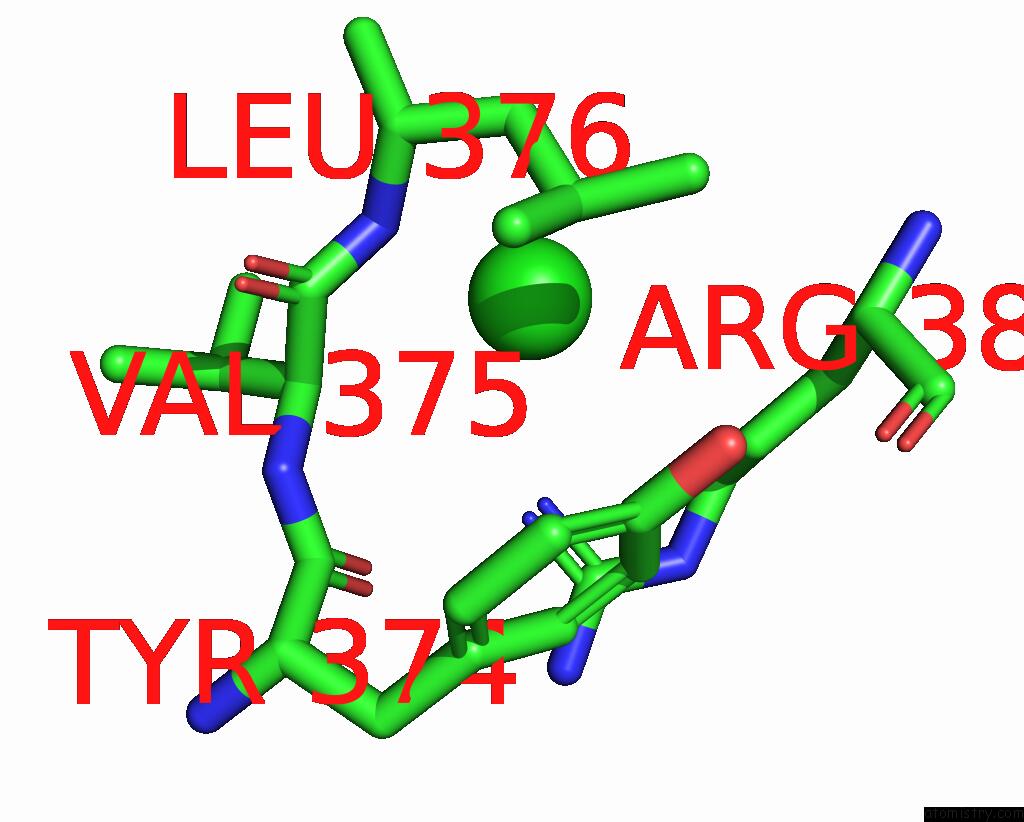
Mono view
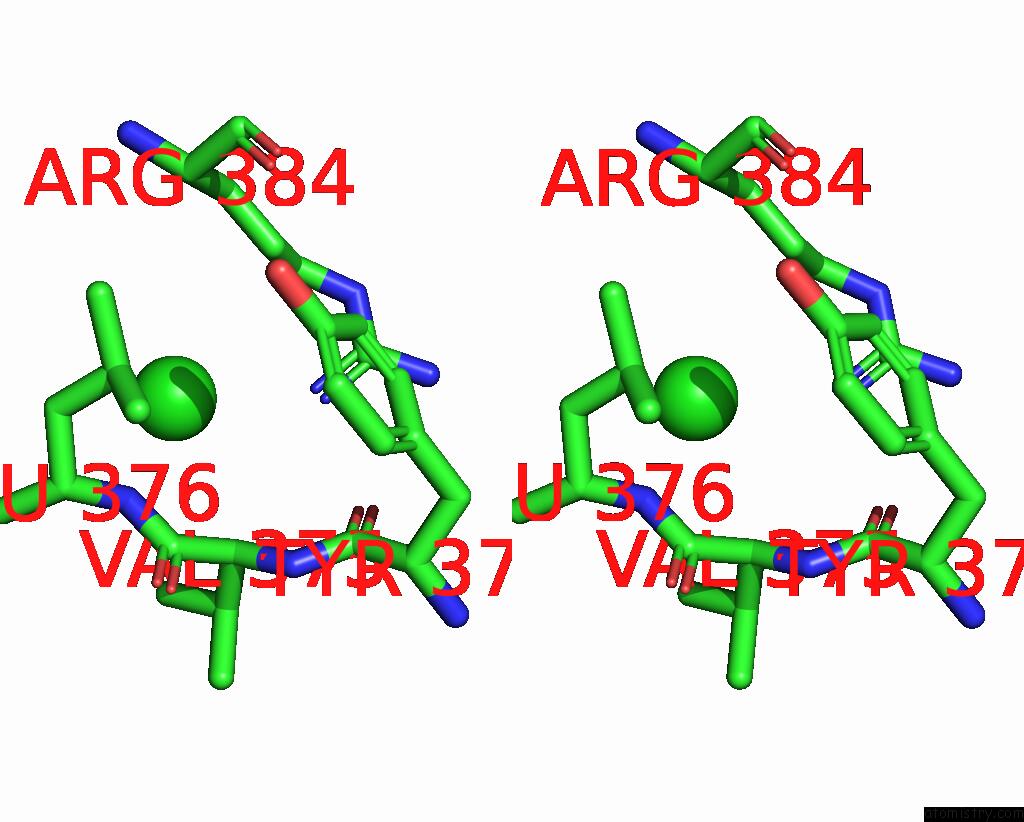
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of the C120G Variant of the Membrane-Bound [Nife]- Hydrogenase From Cupriavidus Necator in the H2-Reduced State at 1.65 A Resolution. within 5.0Å range:
|
Chlorine binding site 3 out of 4 in 8poz
Go back to
Chlorine binding site 3 out
of 4 in the Crystal Structure of the C120G Variant of the Membrane-Bound [Nife]- Hydrogenase From Cupriavidus Necator in the H2-Reduced State at 1.65 A Resolution.
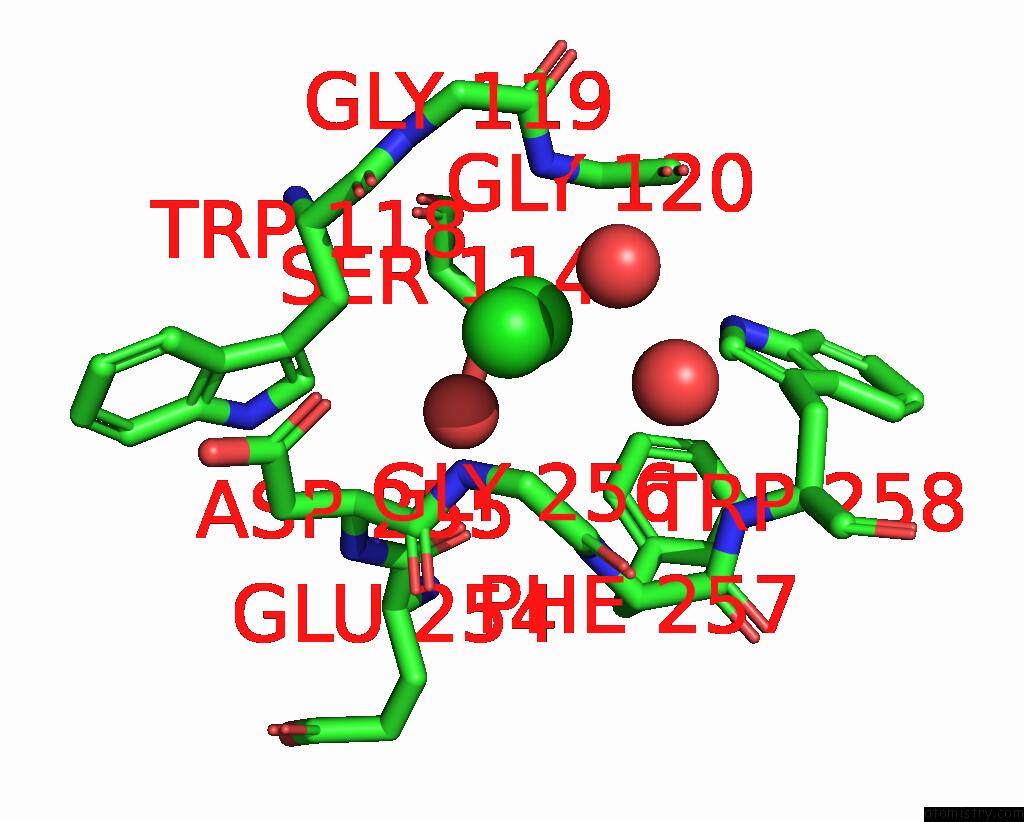
Mono view
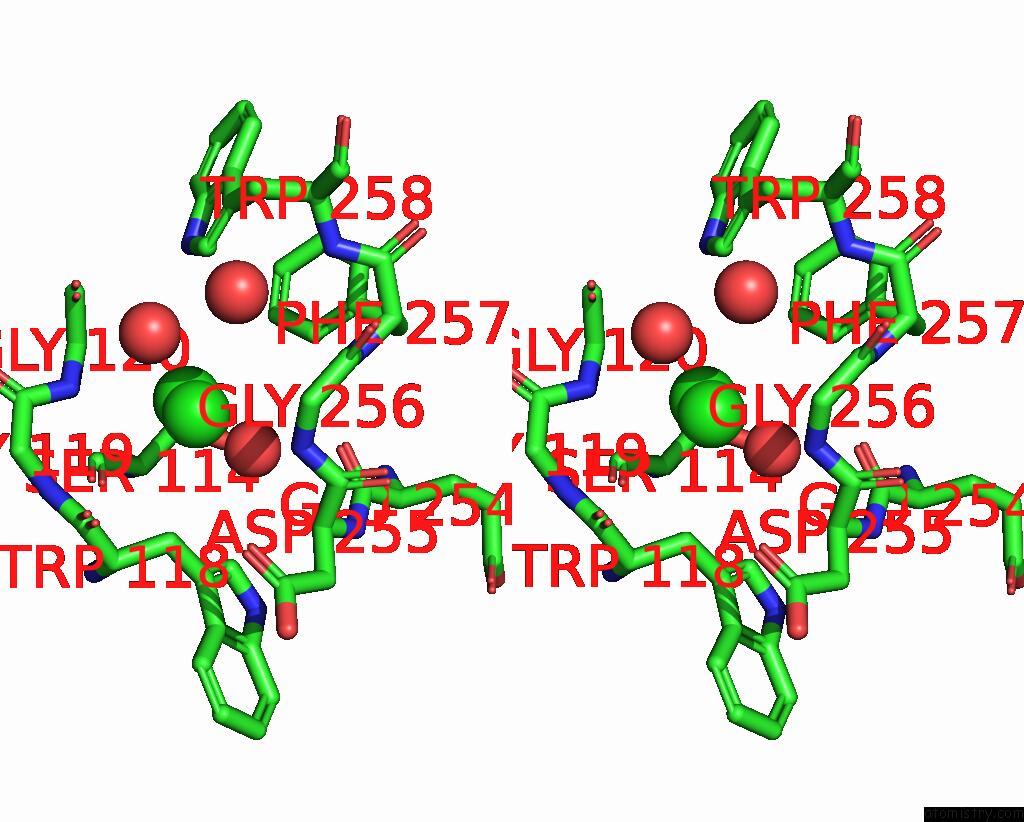
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of the C120G Variant of the Membrane-Bound [Nife]- Hydrogenase From Cupriavidus Necator in the H2-Reduced State at 1.65 A Resolution. within 5.0Å range:
|
Chlorine binding site 4 out of 4 in 8poz
Go back to
Chlorine binding site 4 out
of 4 in the Crystal Structure of the C120G Variant of the Membrane-Bound [Nife]- Hydrogenase From Cupriavidus Necator in the H2-Reduced State at 1.65 A Resolution.
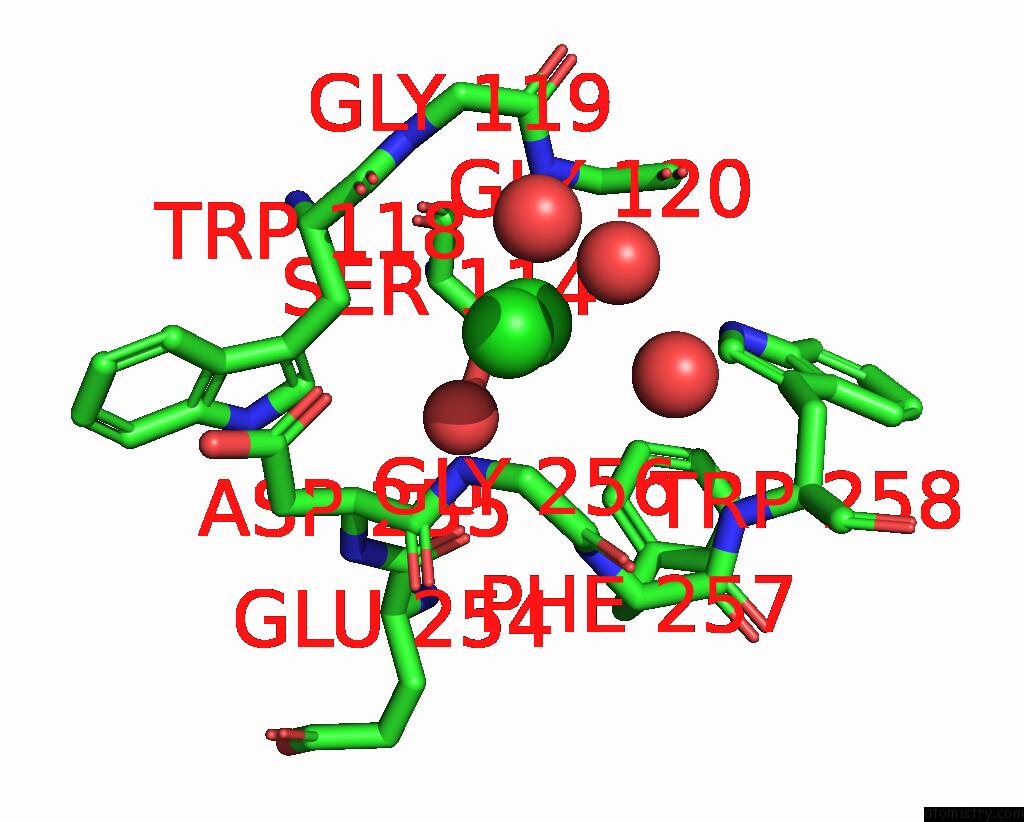
Mono view
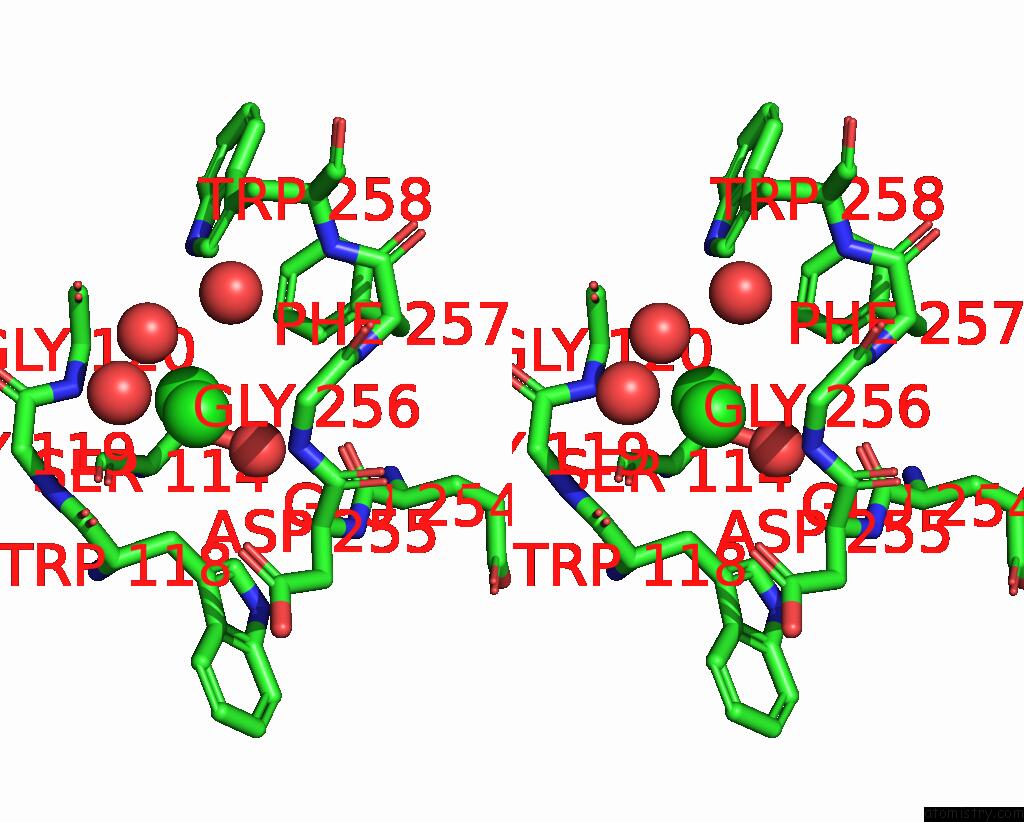
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of the C120G Variant of the Membrane-Bound [Nife]- Hydrogenase From Cupriavidus Necator in the H2-Reduced State at 1.65 A Resolution. within 5.0Å range:
|
Reference:
A.Schmidt,
J.Kalms,
C.Lorent,
S.Katz,
S.Frielingsdorf,
R.M.Evans,
J.Fritsch,
E.Siebert,
C.Teutloff,
F.A.Armstrong,
I.Zebger,
O.Lenz,
P.Scheerer.
Stepwise Conversion of the Cys 6 [4FE-3S] to A Cys 4 [4FE-4S] Cluster and Its Impact on the Oxygen Tolerance of [Nife]-Hydrogenase. Chem Sci V. 14 11105 2023.
ISSN: ISSN 2041-6520
PubMed: 37860641
DOI: 10.1039/D3SC03739H
Page generated: Sun Jul 13 13:13:56 2025
ISSN: ISSN 2041-6520
PubMed: 37860641
DOI: 10.1039/D3SC03739H
Last articles
F in 7P81F in 7PLI
F in 7PJR
F in 7PKV
F in 7PKM
F in 7PKK
F in 7PK8
F in 7PJN
F in 7PK3
F in 7PJC