Chlorine »
PDB 8r5q-8rhq »
8rf4 »
Chlorine in PDB 8rf4: Crystal Structure of N-Terminal Sars-Cov-2 NSP1 in Complex with Fragment Hit 9D4 Refined Against the Anomalous Diffraction Data
Protein crystallography data
The structure of Crystal Structure of N-Terminal Sars-Cov-2 NSP1 in Complex with Fragment Hit 9D4 Refined Against the Anomalous Diffraction Data, PDB code: 8rf4
was solved by
S.Ma,
S.Damfo,
V.Mykhaylyk,
F.Kozielski,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 35.76 / 1.11 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 36.958, 36.958, 141.538, 90, 90, 90 |
| R / Rfree (%) | 18.7 / 21.2 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of N-Terminal Sars-Cov-2 NSP1 in Complex with Fragment Hit 9D4 Refined Against the Anomalous Diffraction Data
(pdb code 8rf4). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Crystal Structure of N-Terminal Sars-Cov-2 NSP1 in Complex with Fragment Hit 9D4 Refined Against the Anomalous Diffraction Data, PDB code: 8rf4:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Crystal Structure of N-Terminal Sars-Cov-2 NSP1 in Complex with Fragment Hit 9D4 Refined Against the Anomalous Diffraction Data, PDB code: 8rf4:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 8rf4
Go back to
Chlorine binding site 1 out
of 2 in the Crystal Structure of N-Terminal Sars-Cov-2 NSP1 in Complex with Fragment Hit 9D4 Refined Against the Anomalous Diffraction Data
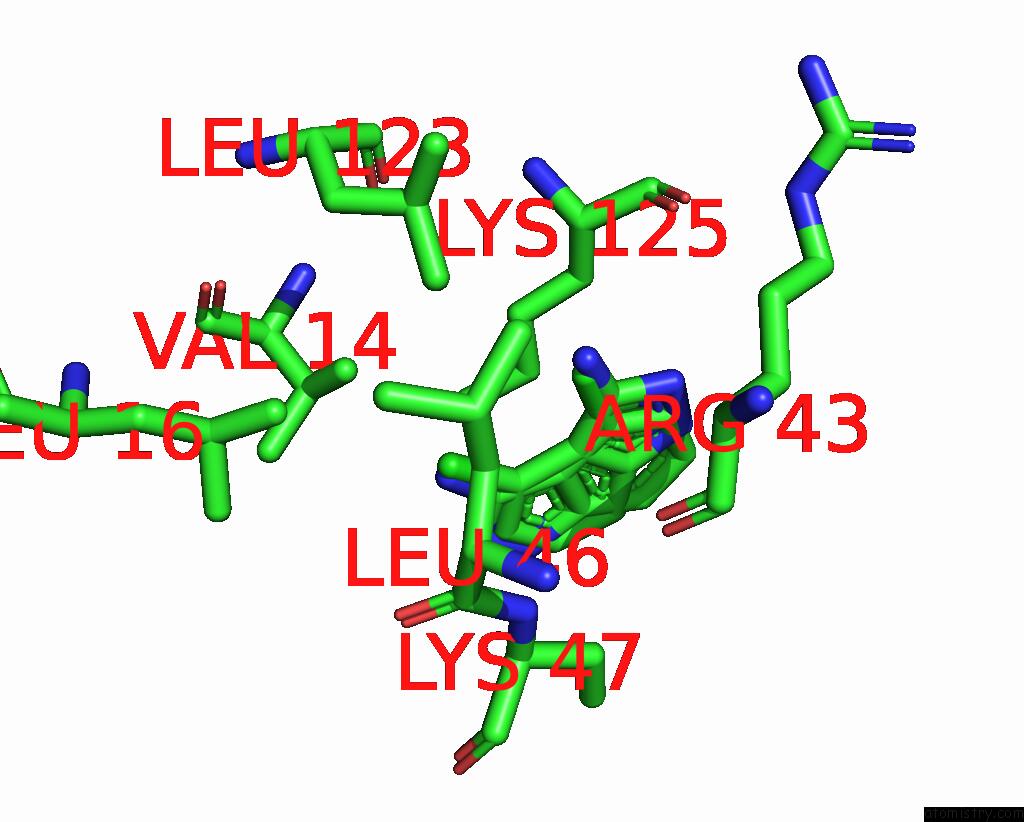
Mono view
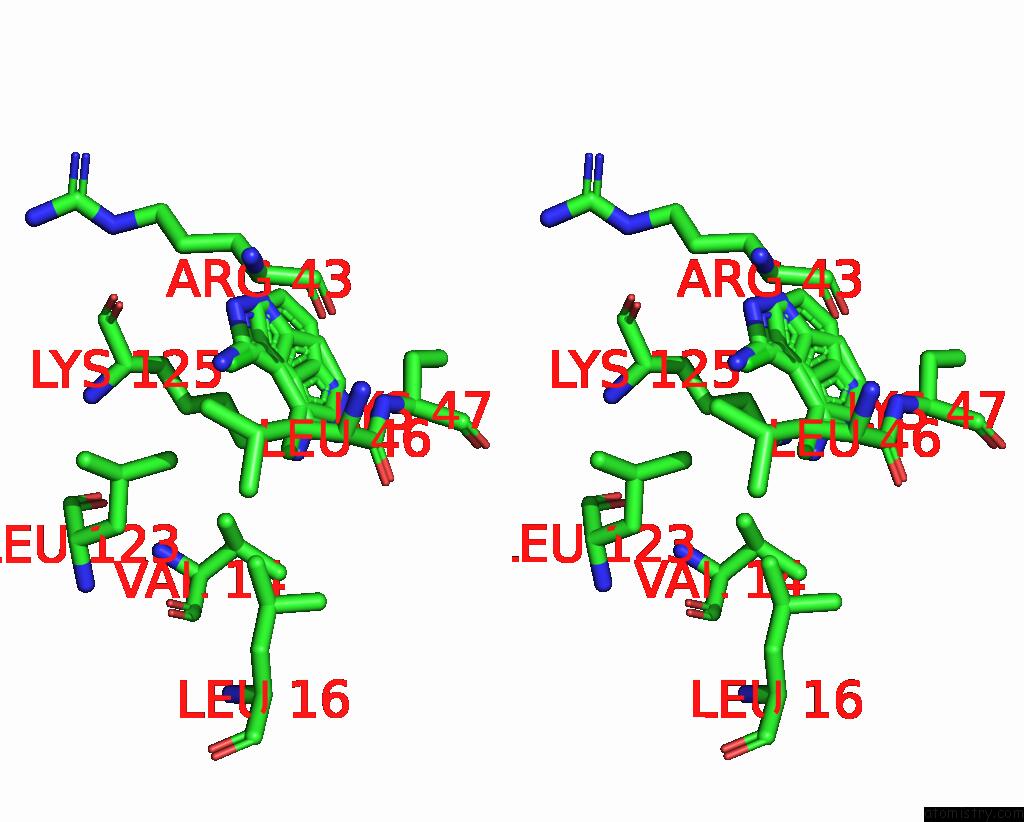
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of N-Terminal Sars-Cov-2 NSP1 in Complex with Fragment Hit 9D4 Refined Against the Anomalous Diffraction Data within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 8rf4
Go back to
Chlorine binding site 2 out
of 2 in the Crystal Structure of N-Terminal Sars-Cov-2 NSP1 in Complex with Fragment Hit 9D4 Refined Against the Anomalous Diffraction Data
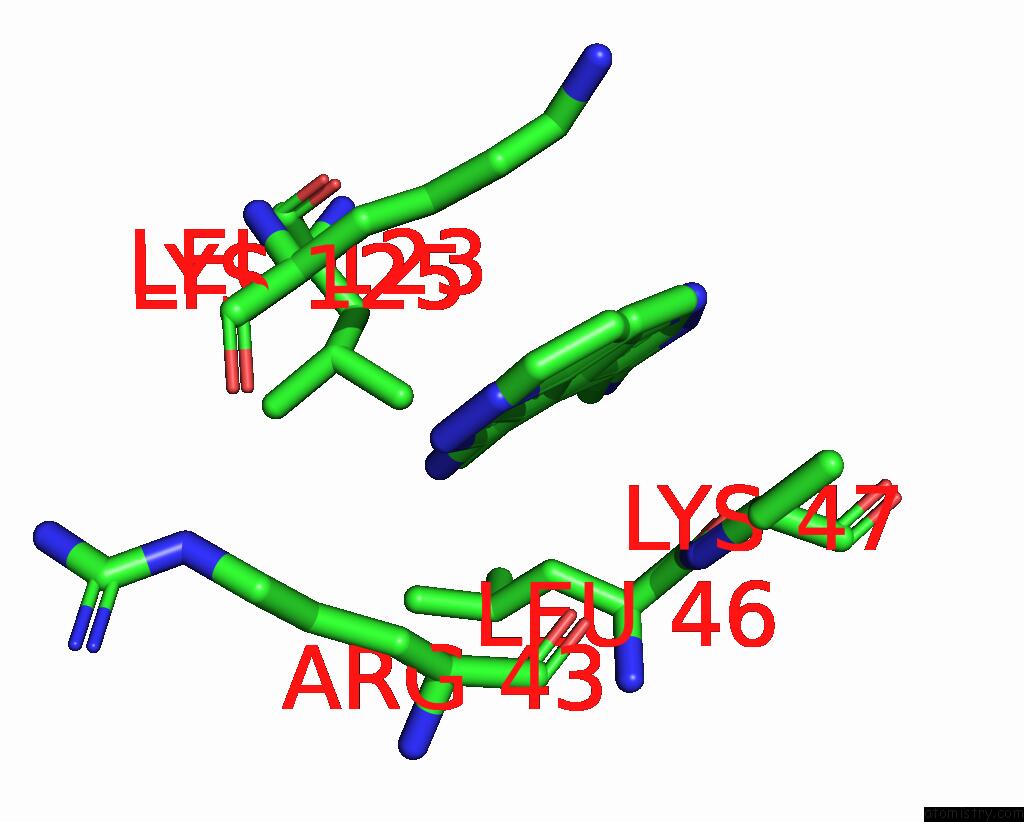
Mono view
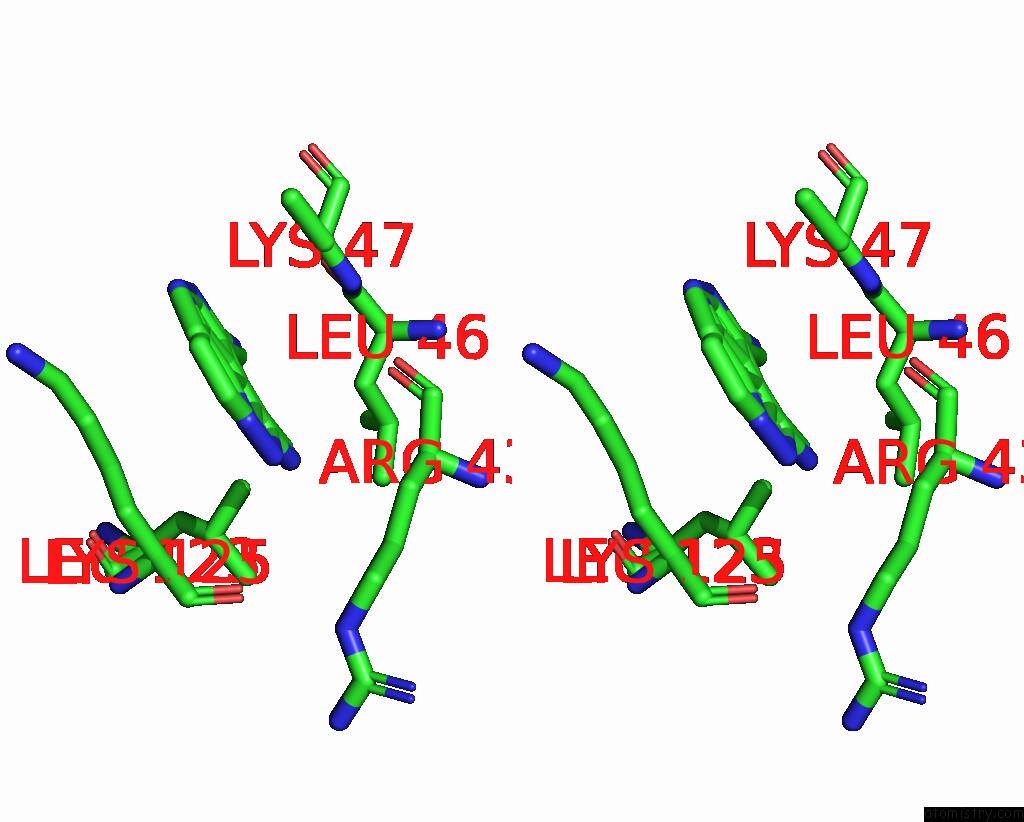
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of N-Terminal Sars-Cov-2 NSP1 in Complex with Fragment Hit 9D4 Refined Against the Anomalous Diffraction Data within 5.0Å range:
|
Reference:
S.Ma,
S.Damfo,
M.W.Bowler,
V.Mykhaylyk,
F.Kozielski.
High-Confidence Placement of Low-Occupancy Fragments Into Electron Density Using the Anomalous Signal of Sulfur and Halogen Atoms. Acta Crystallogr D Struct V. 80 451 2024BIOL.
ISSN: ISSN 2059-7983
PubMed: 38841886
DOI: 10.1107/S2059798324004480
Page generated: Sun Jul 13 13:55:16 2025
ISSN: ISSN 2059-7983
PubMed: 38841886
DOI: 10.1107/S2059798324004480
Last articles
F in 7NTHF in 7NTI
F in 7NPC
F in 7NRG
F in 7NR5
F in 7NQS
F in 7NOS
F in 7NP5
F in 7NDV
F in 7NP6