Chlorine »
PDB 8rwl-8sc9 »
8s1p »
Chlorine in PDB 8s1p: Ylmh Bound to Ptrna-50S
Other elements in 8s1p:
The structure of Ylmh Bound to Ptrna-50S also contains other interesting chemical elements:
| Zinc | (Zn) | 4 atoms |
| Potassium | (K) | 47 atoms |
| Magnesium | (Mg) | 158 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Ylmh Bound to Ptrna-50S
(pdb code 8s1p). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Ylmh Bound to Ptrna-50S, PDB code: 8s1p:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Ylmh Bound to Ptrna-50S, PDB code: 8s1p:
Jump to Chlorine binding site number: 1; 2;
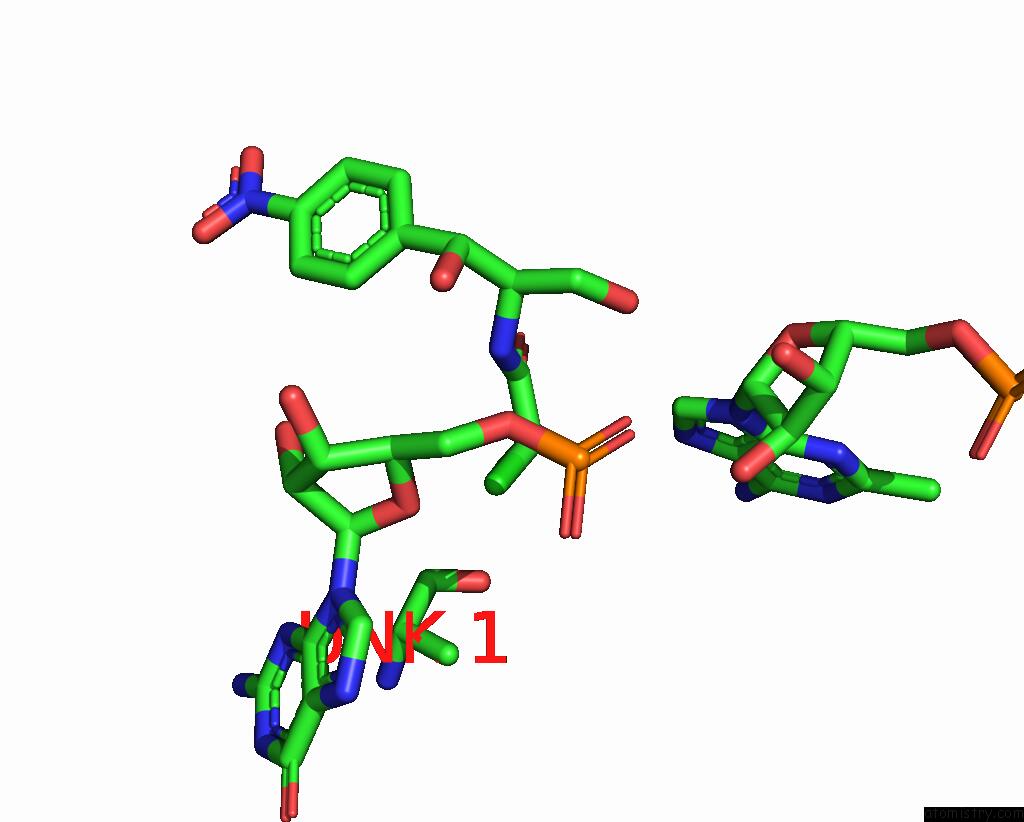
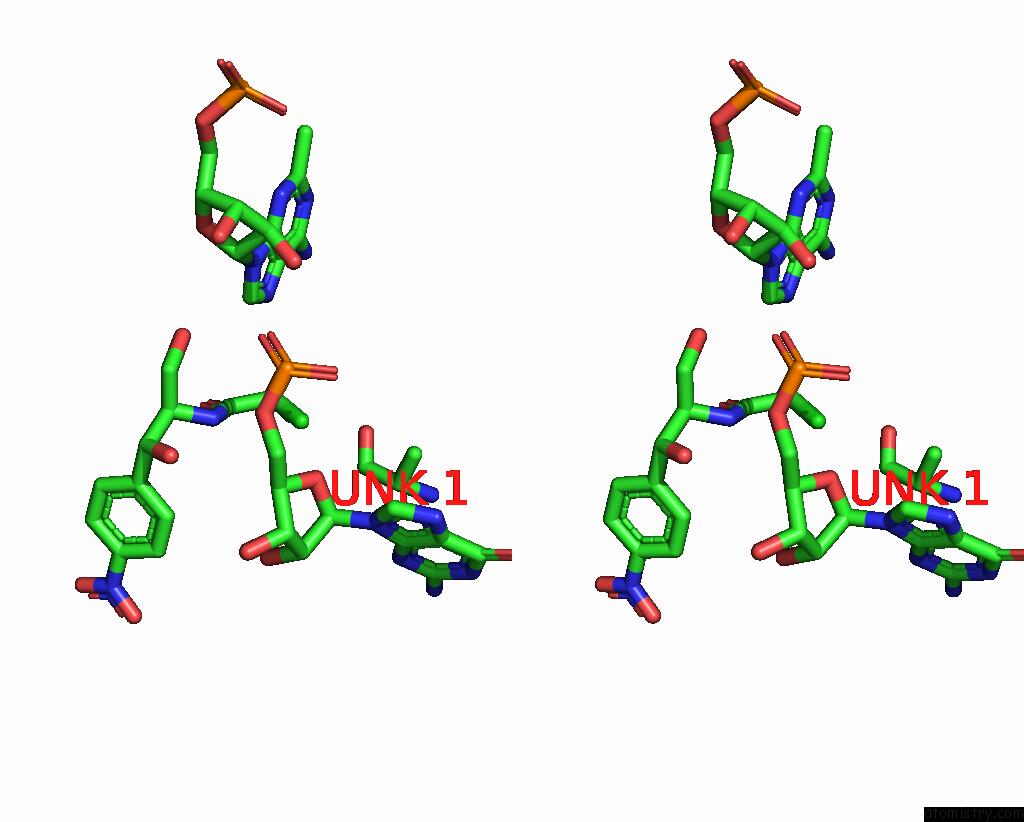
Chlorine binding site 1 out of 2 in 8s1p
Go back to
Chlorine binding site 1 out
of 2 in the Ylmh Bound to Ptrna-50S
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Ylmh Bound to Ptrna-50S within 5.0Å range:
|
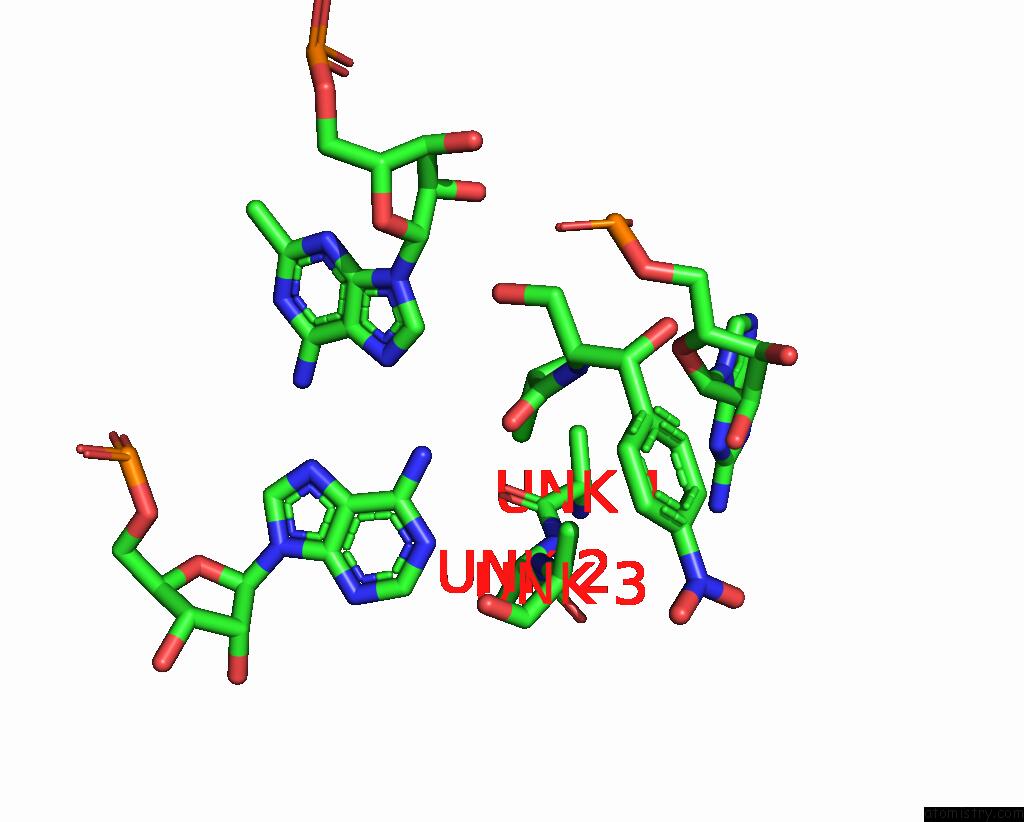
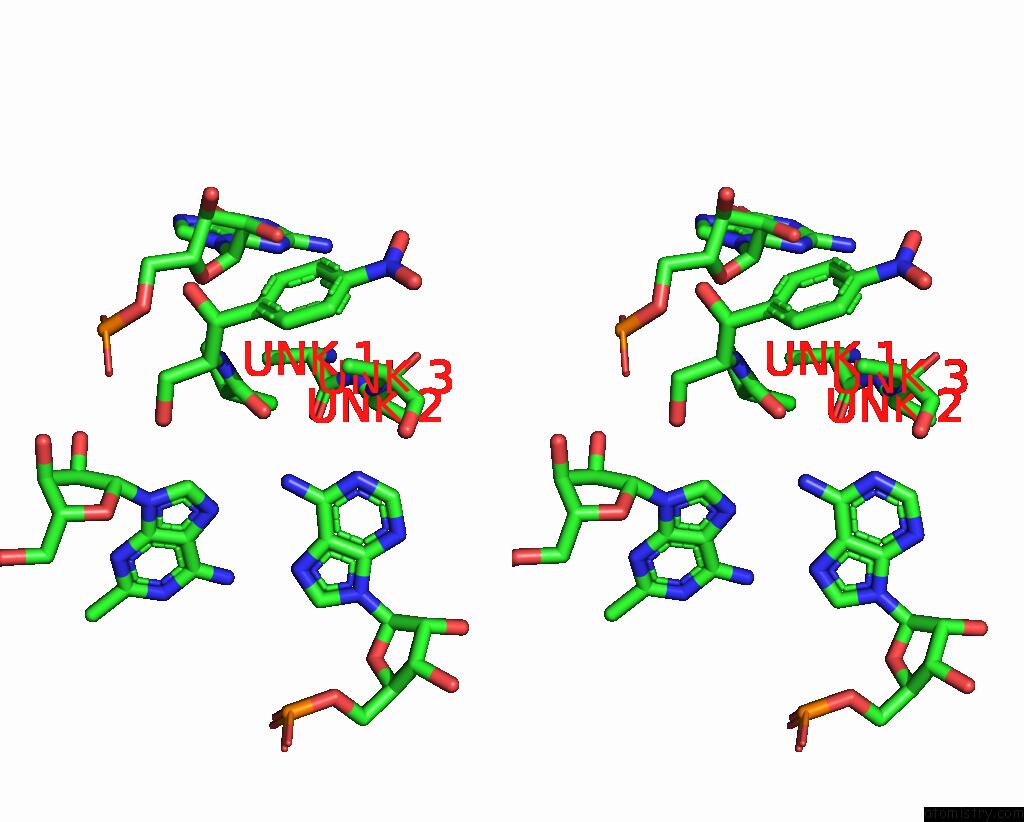
Chlorine binding site 2 out of 2 in 8s1p
Go back to
Chlorine binding site 2 out
of 2 in the Ylmh Bound to Ptrna-50S
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Ylmh Bound to Ptrna-50S within 5.0Å range:
|
Reference:
H.Takada,
H.Paternoga,
K.Fujiwara,
J.A.Nakamoto,
E.N.Park,
L.Dimitrova-Paternoga,
B.Beckert,
M.Saarma,
T.Tenson,
A.R.Buskirk,
G.C.Atkinson,
S.Chiba,
D.N.Wilson,
V.Hauryliuk.
A Role For the S4-Domain Containing Protein Ylmh in Ribosome-Associated Quality Control in Bacillus Subtilis. Nucleic Acids Res. 2024.
ISSN: ESSN 1362-4962
PubMed: 38811035
DOI: 10.1093/NAR/GKAE399
Page generated: Sun Jul 13 14:02:53 2025
ISSN: ESSN 1362-4962
PubMed: 38811035
DOI: 10.1093/NAR/GKAE399
Last articles
F in 7MPBF in 7MR6
F in 7MNG
F in 7MR5
F in 7MON
F in 7MOG
F in 7MOO
F in 7MML
F in 7MMI
F in 7MMK