Chlorine »
PDB 8rwl-8sc9 »
8s4p »
Chlorine in PDB 8s4p: Crystal Structure of An Ene-Reductase From Penicillium Steckii
Protein crystallography data
The structure of Crystal Structure of An Ene-Reductase From Penicillium Steckii, PDB code: 8s4p
was solved by
H.J.Rozeboom,
M.W.Fraaije,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 86.88 / 2.29 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 107.482, 175.292, 107.532, 90, 111.53, 90 |
| R / Rfree (%) | 20.1 / 27.5 |
Other elements in 8s4p:
The structure of Crystal Structure of An Ene-Reductase From Penicillium Steckii also contains other interesting chemical elements:
| Calcium | (Ca) | 6 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of An Ene-Reductase From Penicillium Steckii
(pdb code 8s4p). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 7 binding sites of Chlorine where determined in the Crystal Structure of An Ene-Reductase From Penicillium Steckii, PDB code: 8s4p:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7;
In total 7 binding sites of Chlorine where determined in the Crystal Structure of An Ene-Reductase From Penicillium Steckii, PDB code: 8s4p:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7;
Chlorine binding site 1 out of 7 in 8s4p
Go back to
Chlorine binding site 1 out
of 7 in the Crystal Structure of An Ene-Reductase From Penicillium Steckii
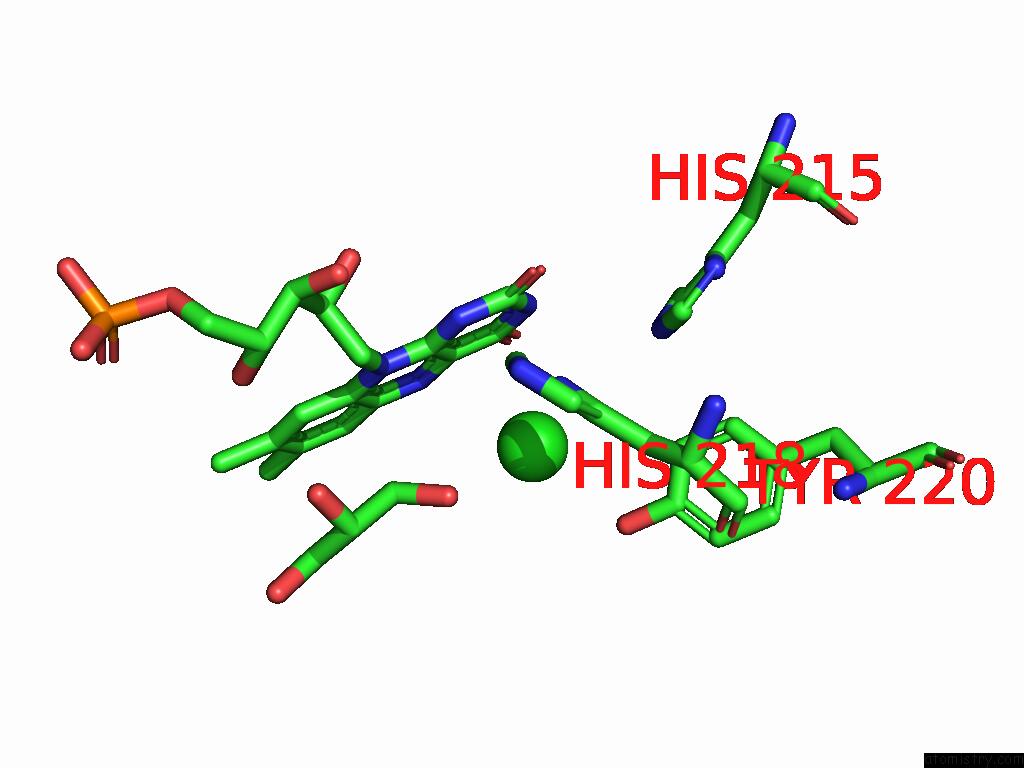
Mono view
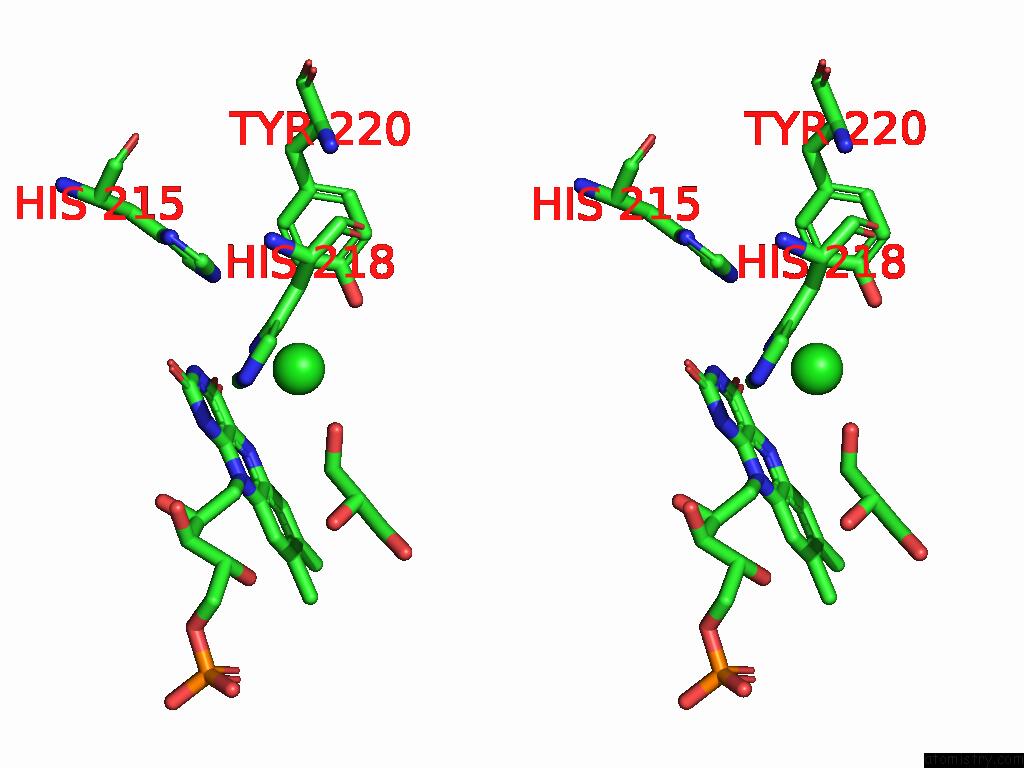
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of An Ene-Reductase From Penicillium Steckii within 5.0Å range:
|
Chlorine binding site 2 out of 7 in 8s4p
Go back to
Chlorine binding site 2 out
of 7 in the Crystal Structure of An Ene-Reductase From Penicillium Steckii
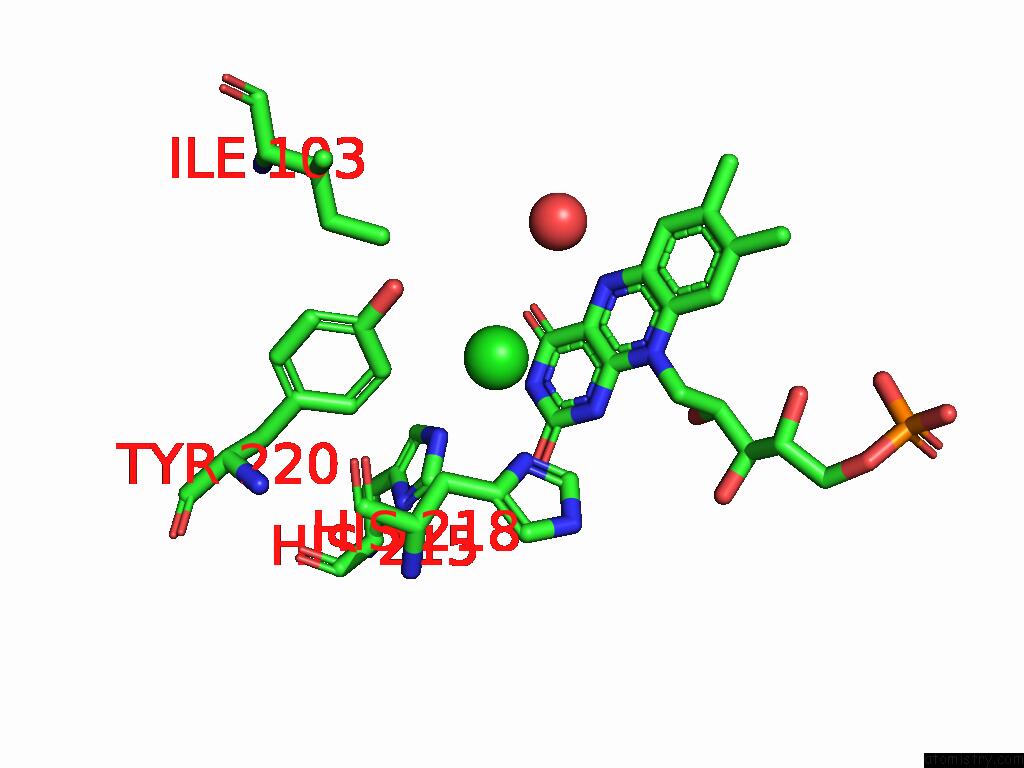
Mono view
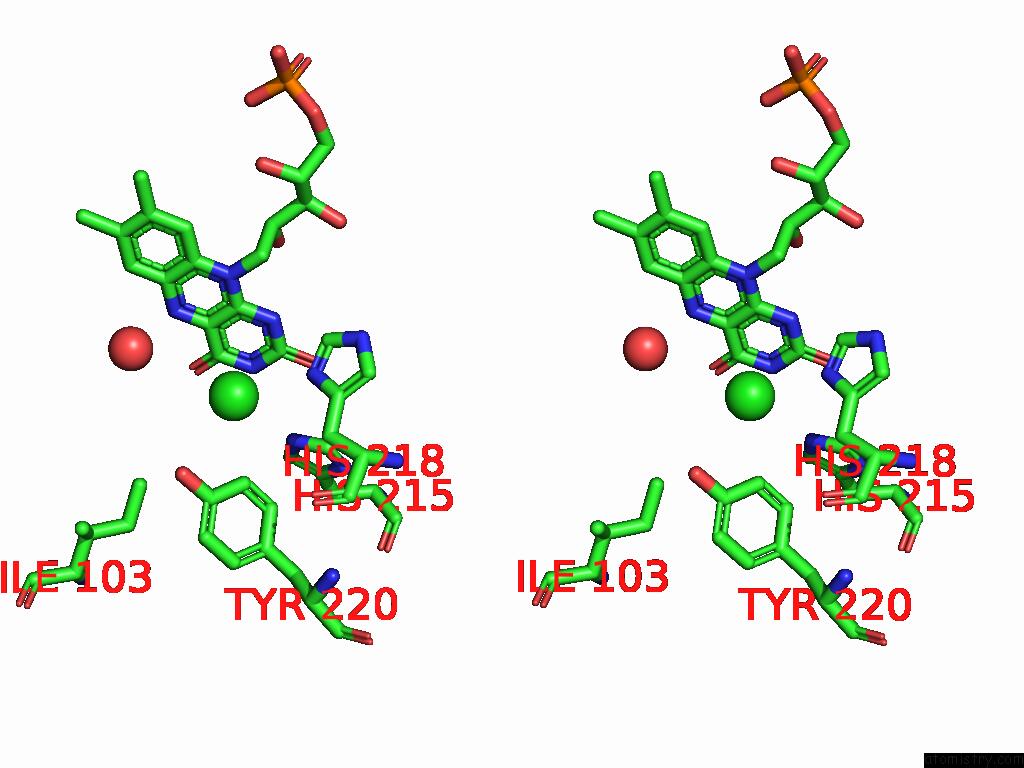
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of An Ene-Reductase From Penicillium Steckii within 5.0Å range:
|
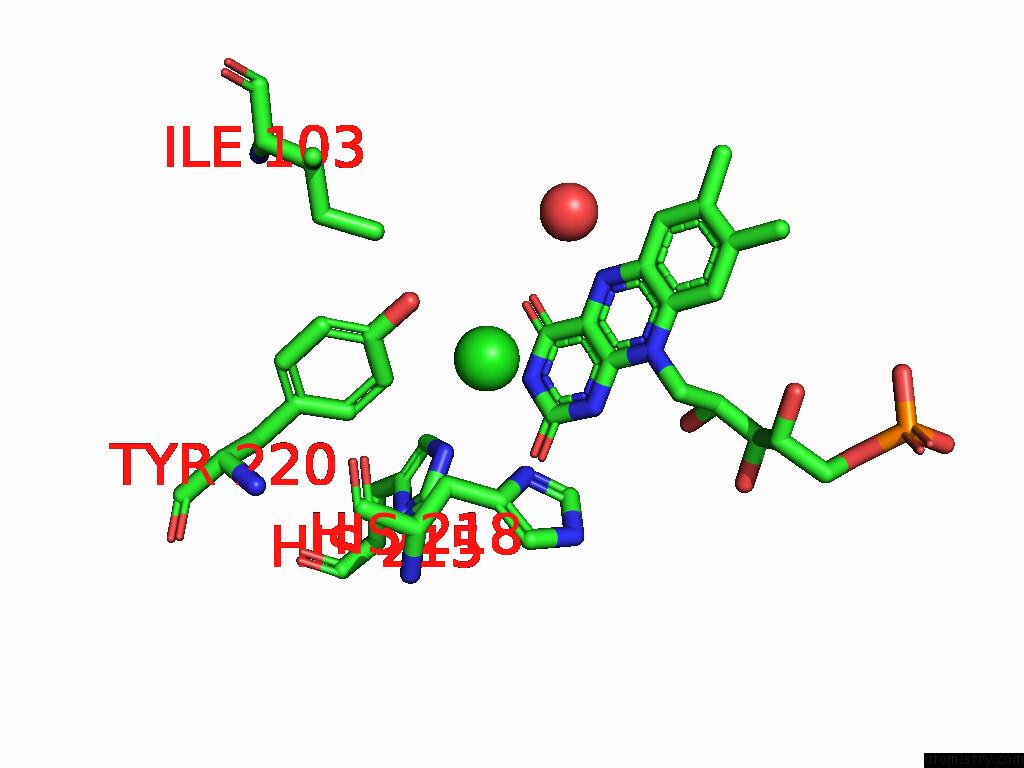
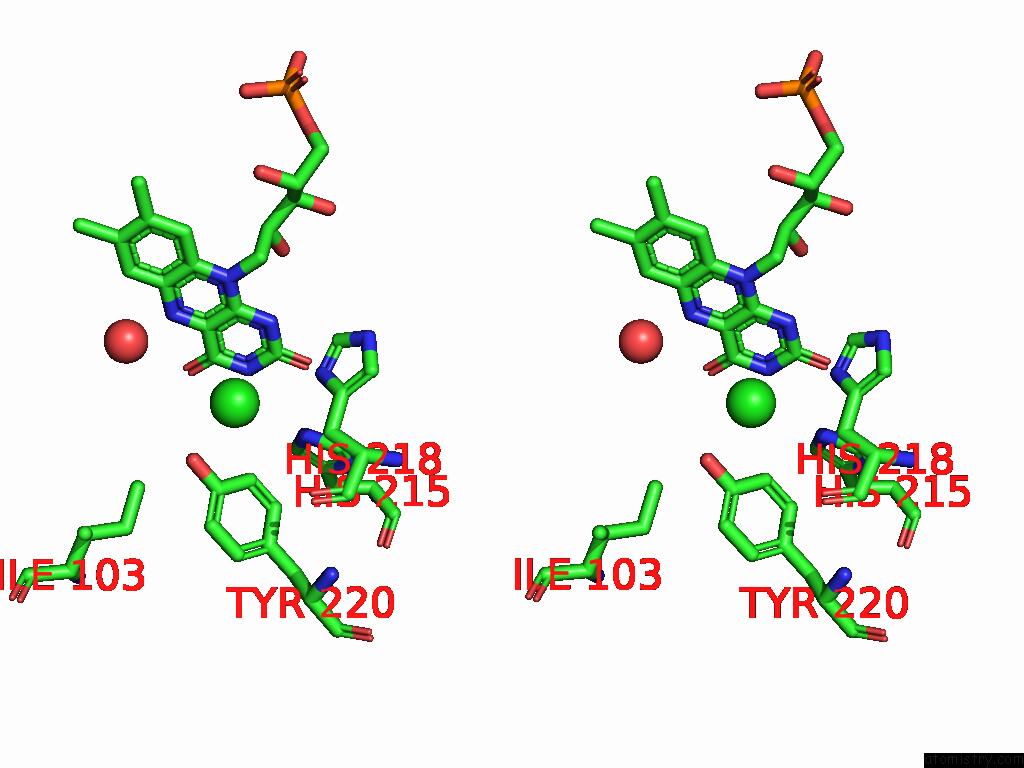
Chlorine binding site 3 out of 7 in 8s4p
Go back to
Chlorine binding site 3 out
of 7 in the Crystal Structure of An Ene-Reductase From Penicillium Steckii
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of An Ene-Reductase From Penicillium Steckii within 5.0Å range:
|
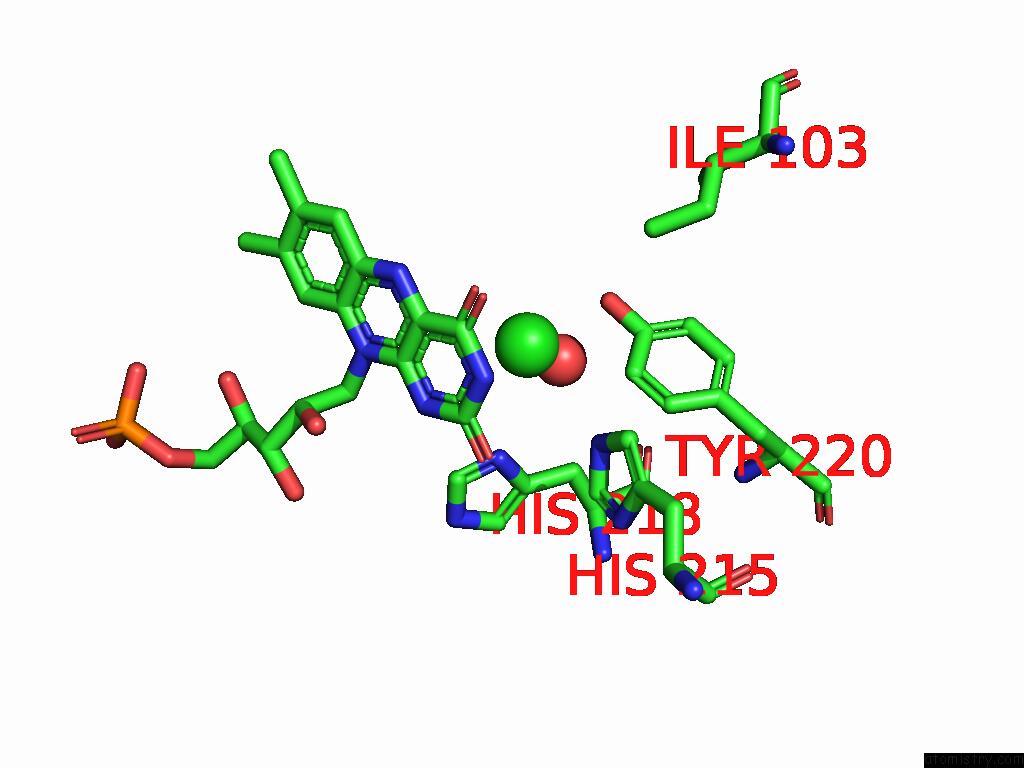
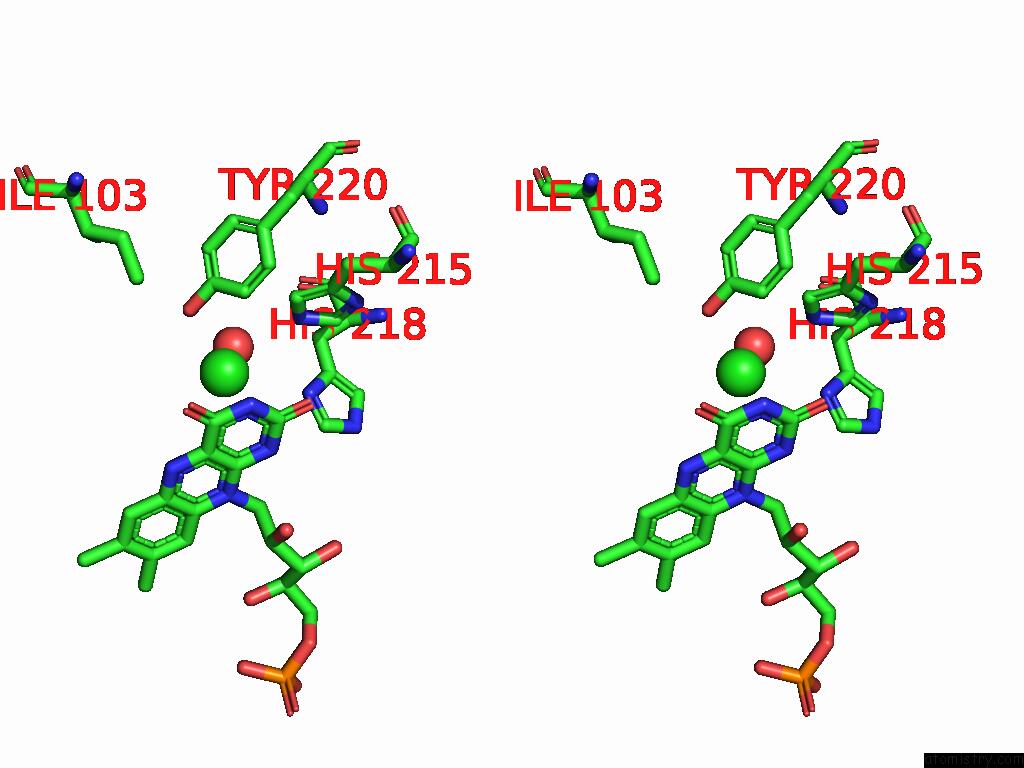
Chlorine binding site 4 out of 7 in 8s4p
Go back to
Chlorine binding site 4 out
of 7 in the Crystal Structure of An Ene-Reductase From Penicillium Steckii
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of An Ene-Reductase From Penicillium Steckii within 5.0Å range:
|
Chlorine binding site 5 out of 7 in 8s4p
Go back to
Chlorine binding site 5 out
of 7 in the Crystal Structure of An Ene-Reductase From Penicillium Steckii
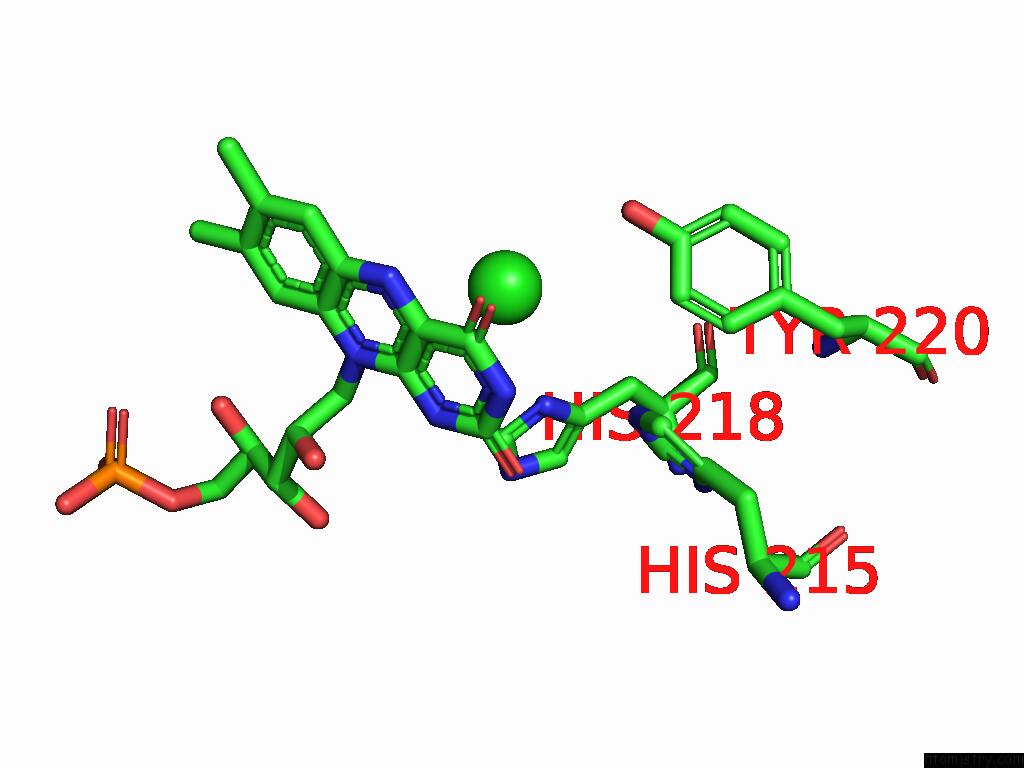
Mono view
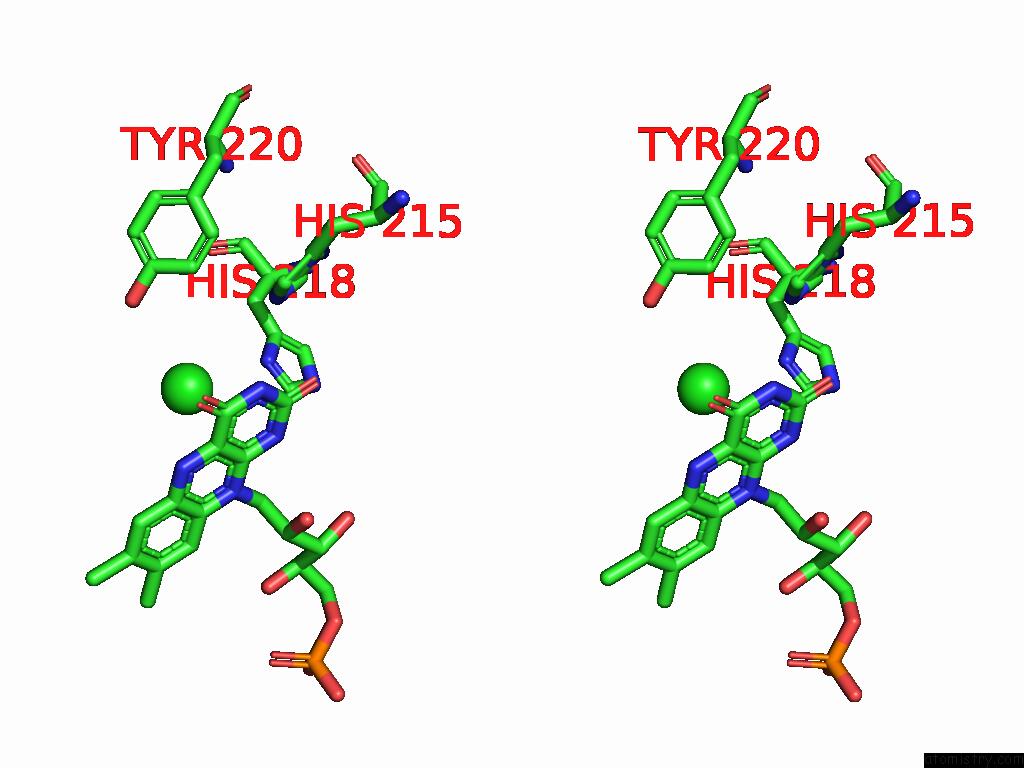
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Crystal Structure of An Ene-Reductase From Penicillium Steckii within 5.0Å range:
|
Chlorine binding site 6 out of 7 in 8s4p
Go back to
Chlorine binding site 6 out
of 7 in the Crystal Structure of An Ene-Reductase From Penicillium Steckii
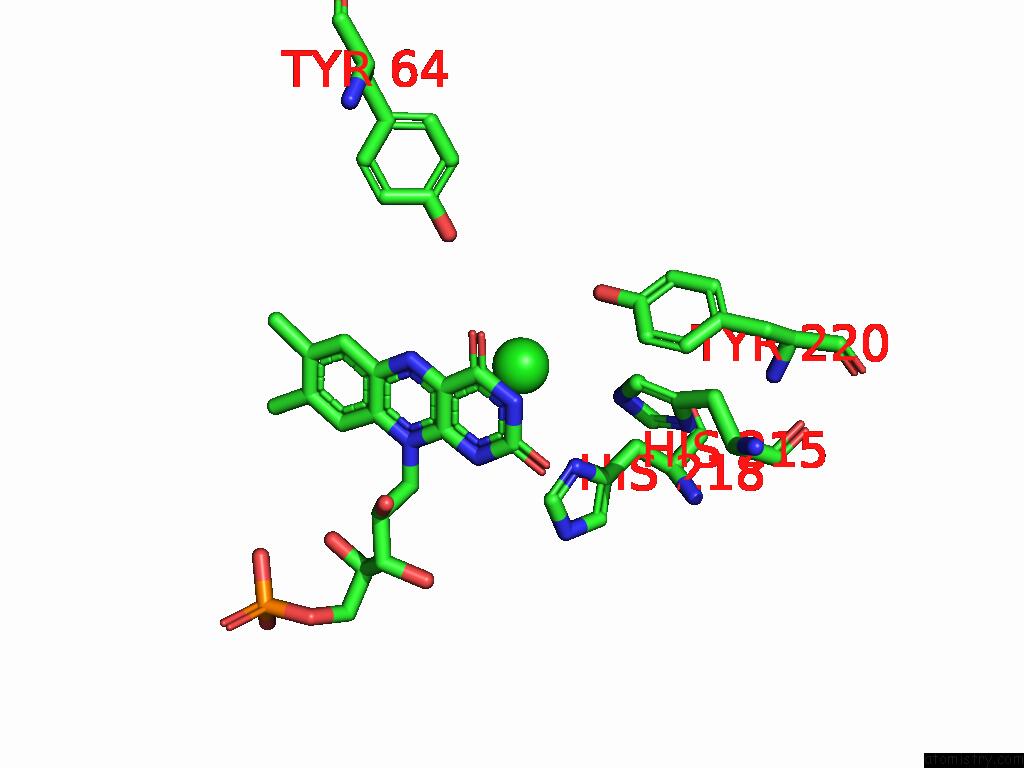
Mono view
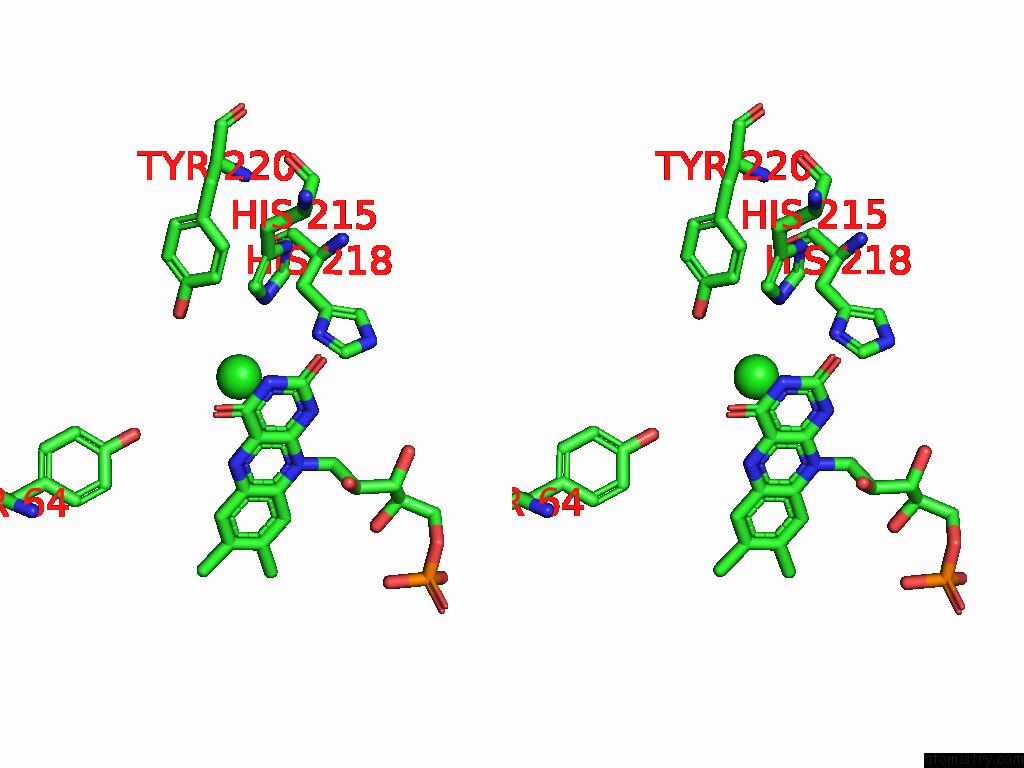
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Crystal Structure of An Ene-Reductase From Penicillium Steckii within 5.0Å range:
|
Chlorine binding site 7 out of 7 in 8s4p
Go back to
Chlorine binding site 7 out
of 7 in the Crystal Structure of An Ene-Reductase From Penicillium Steckii
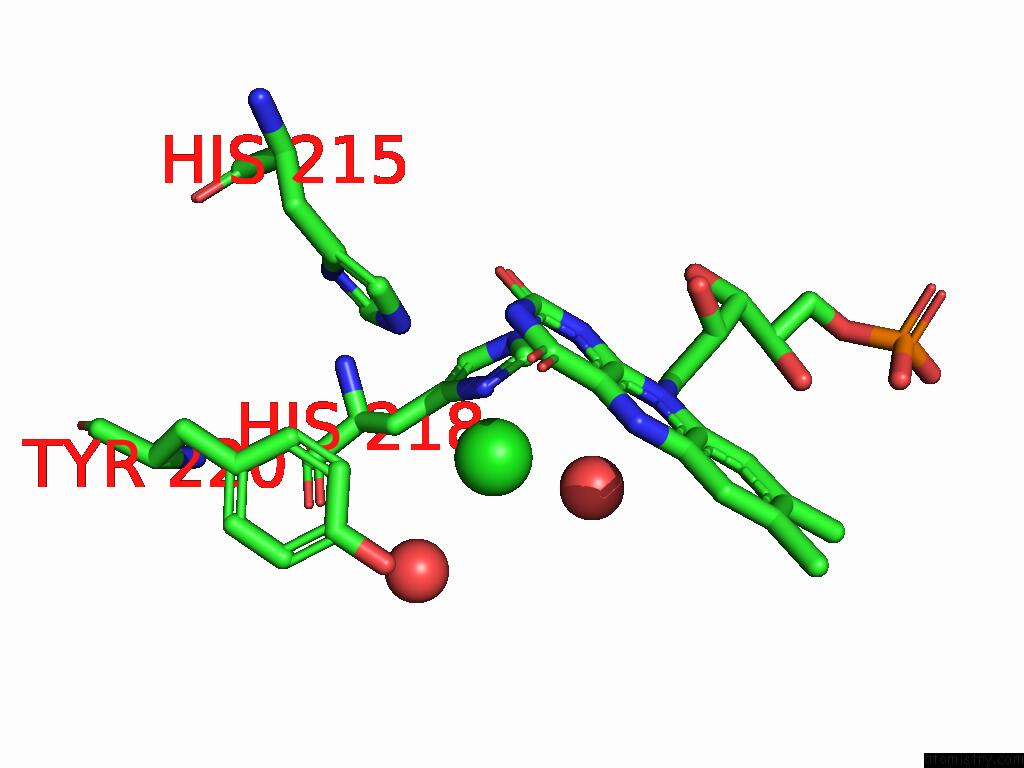
Mono view
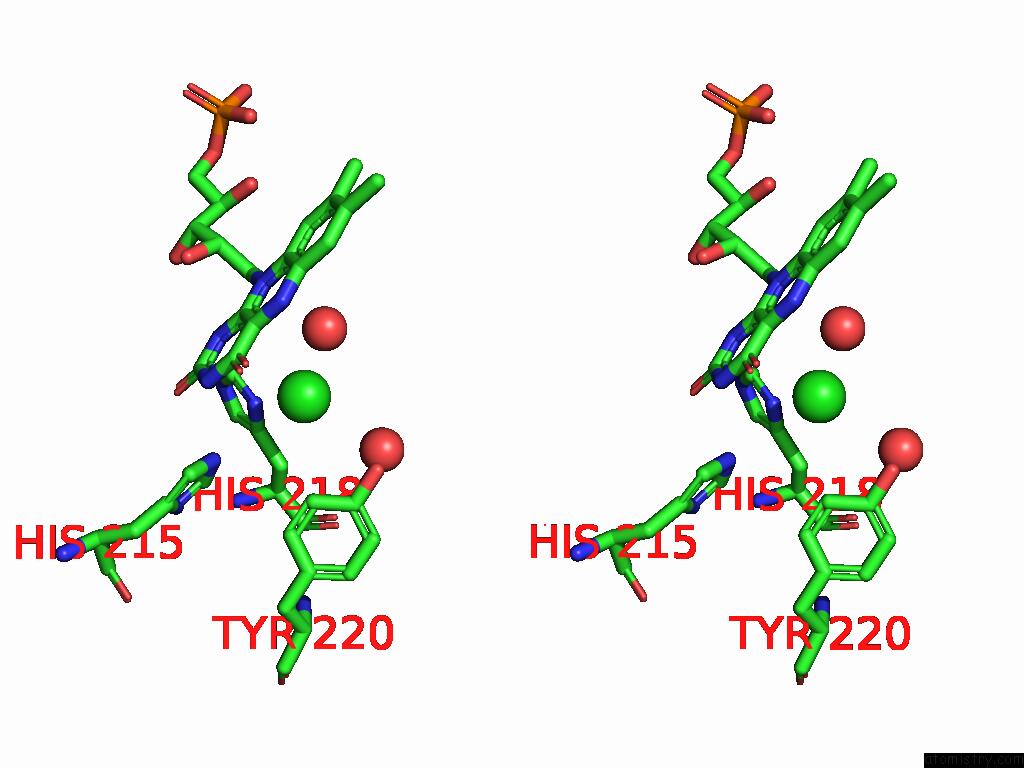
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 7 of Crystal Structure of An Ene-Reductase From Penicillium Steckii within 5.0Å range:
|
Reference:
P.H.Damada,
H.J.Rozeboom,
A.L.M.Porto,
M.W.Fraaije.
Obtaining and Characterization of Six Novel Ene-Reductases Form Different Penicillium Steckii Strais To Be Published.
Page generated: Sun Jul 13 14:03:24 2025
Last articles
Co in 6VV9Co in 6VV8
Co in 6VV7
Co in 6VV6
Co in 6VSG
Co in 6VSF
Co in 6VMY
Co in 6VSE
Co in 6VSD
Co in 6VS9