Chlorine »
PDB 8sqq-8t3p »
8ssj »
Chlorine in PDB 8ssj: Room-Temperature X-Ray Structure of Human Mitochondrial Serine Hydroxymethyltransferase (HSHMT2)
Enzymatic activity of Room-Temperature X-Ray Structure of Human Mitochondrial Serine Hydroxymethyltransferase (HSHMT2)
All present enzymatic activity of Room-Temperature X-Ray Structure of Human Mitochondrial Serine Hydroxymethyltransferase (HSHMT2):
2.1.2.1;
2.1.2.1;
Protein crystallography data
The structure of Room-Temperature X-Ray Structure of Human Mitochondrial Serine Hydroxymethyltransferase (HSHMT2), PDB code: 8ssj
was solved by
V.N.Drago,
A.Kovalevsky,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 46.65 / 2.50 |
| Space group | P 65 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 161.609, 161.609, 210.784, 90, 90, 120 |
| R / Rfree (%) | 17.2 / 20.5 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Room-Temperature X-Ray Structure of Human Mitochondrial Serine Hydroxymethyltransferase (HSHMT2)
(pdb code 8ssj). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Room-Temperature X-Ray Structure of Human Mitochondrial Serine Hydroxymethyltransferase (HSHMT2), PDB code: 8ssj:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Room-Temperature X-Ray Structure of Human Mitochondrial Serine Hydroxymethyltransferase (HSHMT2), PDB code: 8ssj:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 8ssj
Go back to
Chlorine binding site 1 out
of 2 in the Room-Temperature X-Ray Structure of Human Mitochondrial Serine Hydroxymethyltransferase (HSHMT2)

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Room-Temperature X-Ray Structure of Human Mitochondrial Serine Hydroxymethyltransferase (HSHMT2) within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 8ssj
Go back to
Chlorine binding site 2 out
of 2 in the Room-Temperature X-Ray Structure of Human Mitochondrial Serine Hydroxymethyltransferase (HSHMT2)
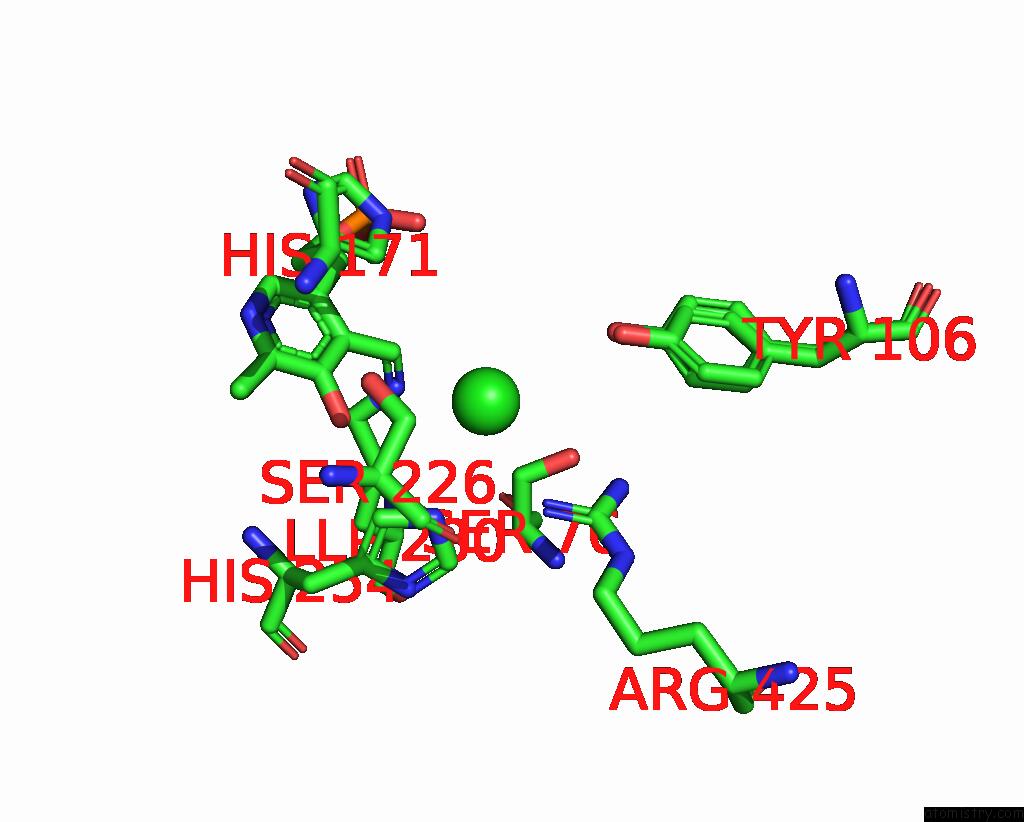
Mono view
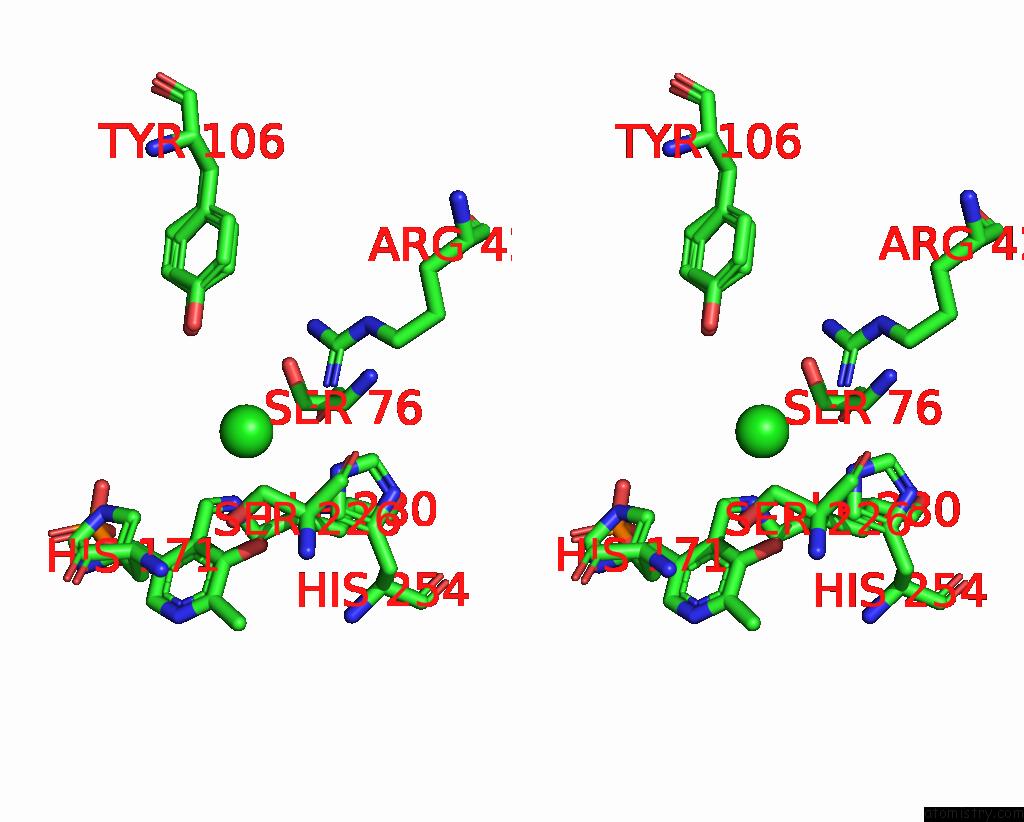
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Room-Temperature X-Ray Structure of Human Mitochondrial Serine Hydroxymethyltransferase (HSHMT2) within 5.0Å range:
|
Reference:
V.N.Drago,
C.Campos,
M.Hooper,
A.Collins,
O.Gerlits,
K.L.Weiss,
M.P.Blakeley,
R.S.Phillips,
A.Kovalevsky.
Revealing Protonation States and Tracking Substrate in Serine Hydroxymethyltransferase with Room-Temperature X-Ray and Neutron Crystallography. Commun Chem V. 6 162 2023.
ISSN: ESSN 2399-3669
PubMed: 37532884
DOI: 10.1038/S42004-023-00964-9
Page generated: Sun Jul 13 14:16:39 2025
ISSN: ESSN 2399-3669
PubMed: 37532884
DOI: 10.1038/S42004-023-00964-9
Last articles
F in 7MPBF in 7MR6
F in 7MNG
F in 7MR5
F in 7MON
F in 7MOG
F in 7MOO
F in 7MML
F in 7MMI
F in 7MMK