Chlorine »
PDB 8tg4-8tsz »
8tr6 »
Chlorine in PDB 8tr6: Cryo-Em Structure of the Rat P2X7 Receptor in Complex with the Allosteric Antagonist A438079
Other elements in 8tr6:
The structure of Cryo-Em Structure of the Rat P2X7 Receptor in Complex with the Allosteric Antagonist A438079 also contains other interesting chemical elements:
| Zinc | (Zn) | 6 atoms |
| Sodium | (Na) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Cryo-Em Structure of the Rat P2X7 Receptor in Complex with the Allosteric Antagonist A438079
(pdb code 8tr6). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 6 binding sites of Chlorine where determined in the Cryo-Em Structure of the Rat P2X7 Receptor in Complex with the Allosteric Antagonist A438079, PDB code: 8tr6:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Chlorine where determined in the Cryo-Em Structure of the Rat P2X7 Receptor in Complex with the Allosteric Antagonist A438079, PDB code: 8tr6:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
Chlorine binding site 1 out of 6 in 8tr6
Go back to
Chlorine binding site 1 out
of 6 in the Cryo-Em Structure of the Rat P2X7 Receptor in Complex with the Allosteric Antagonist A438079
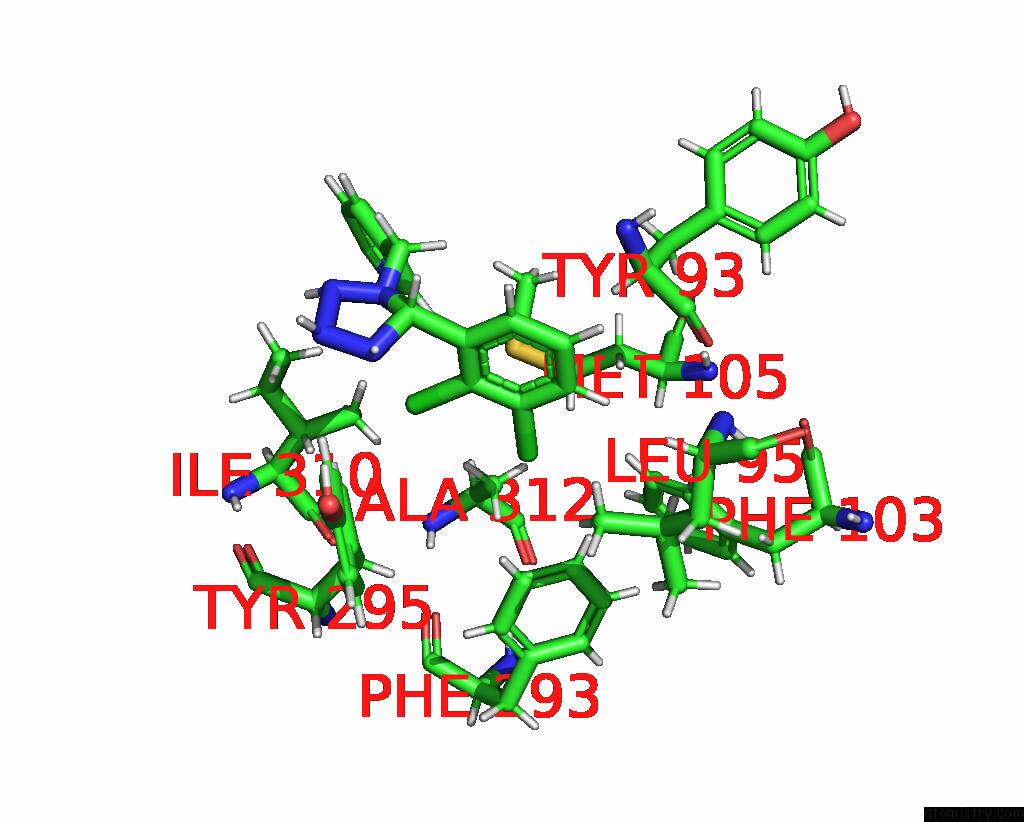
Mono view
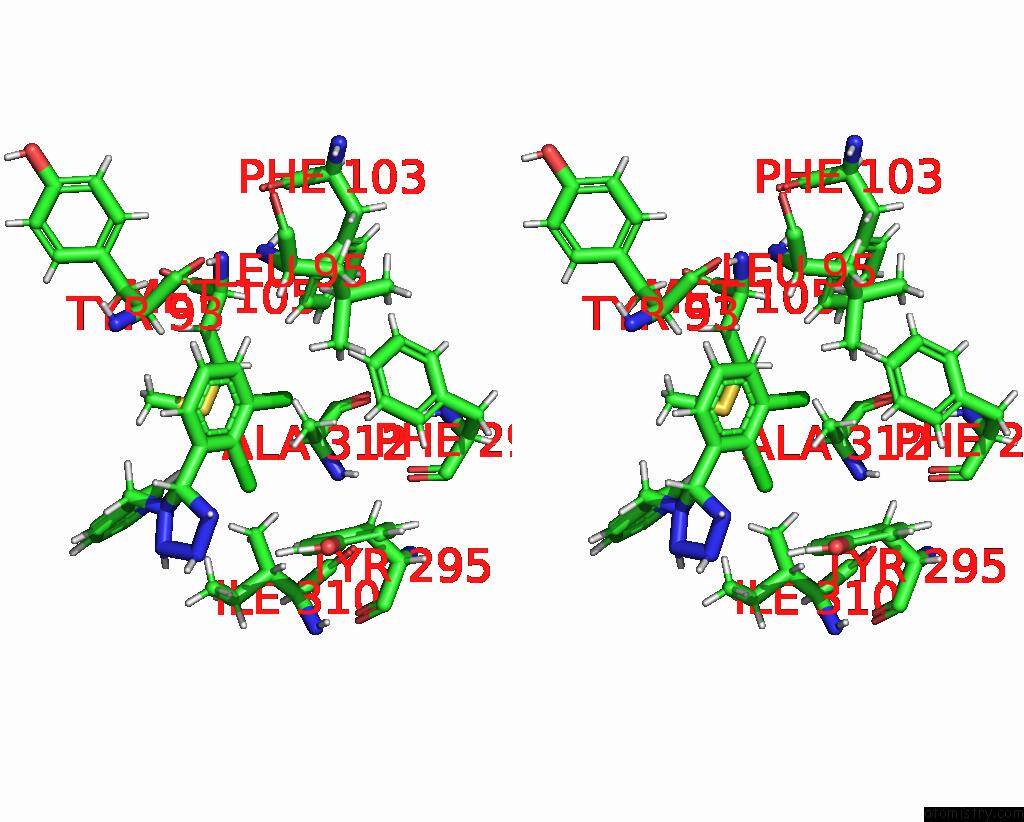
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Cryo-Em Structure of the Rat P2X7 Receptor in Complex with the Allosteric Antagonist A438079 within 5.0Å range:
|
Chlorine binding site 2 out of 6 in 8tr6
Go back to
Chlorine binding site 2 out
of 6 in the Cryo-Em Structure of the Rat P2X7 Receptor in Complex with the Allosteric Antagonist A438079
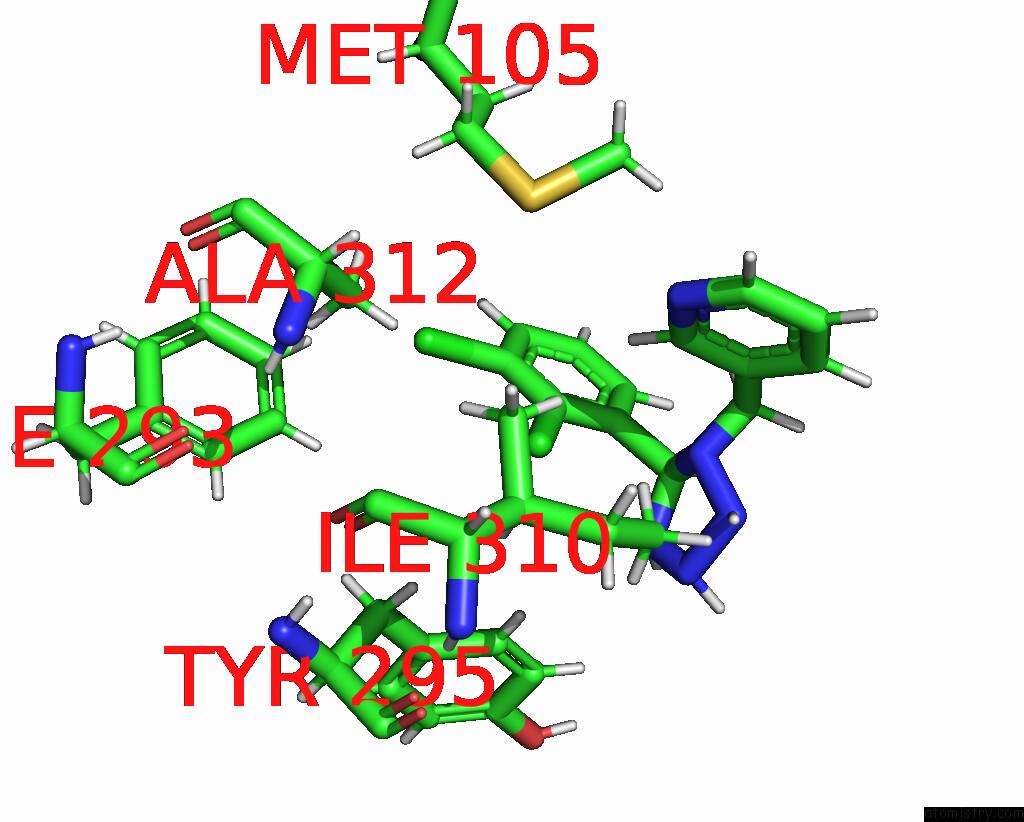
Mono view
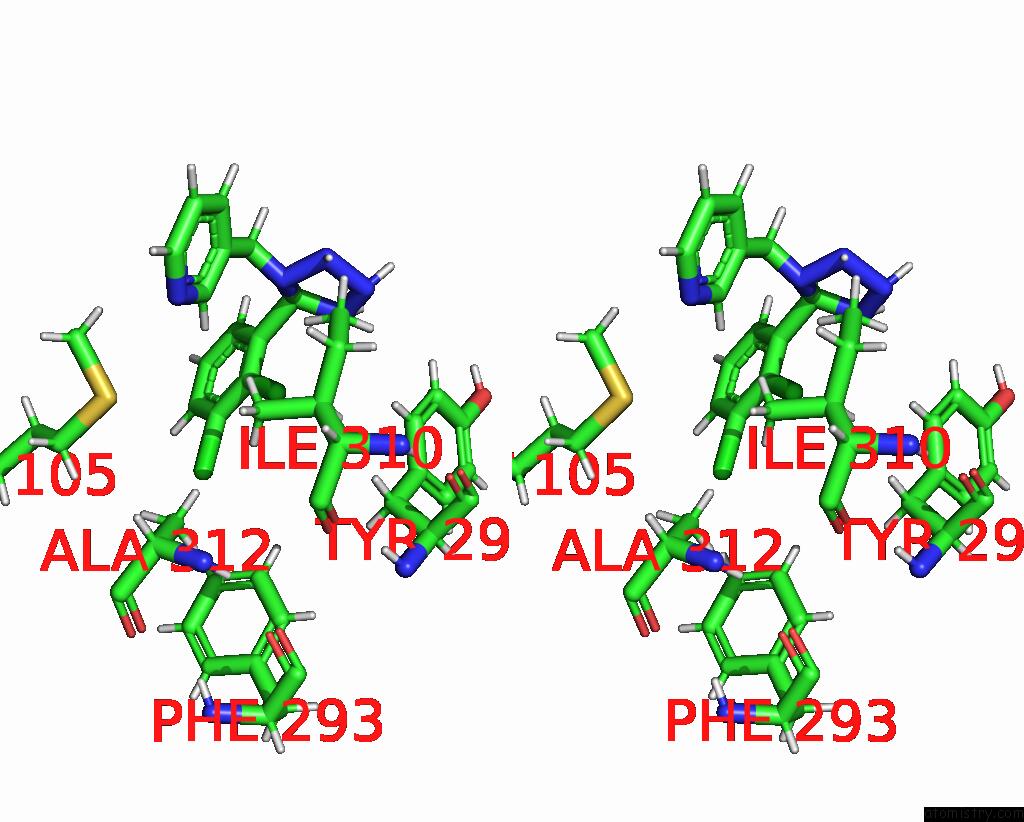
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Cryo-Em Structure of the Rat P2X7 Receptor in Complex with the Allosteric Antagonist A438079 within 5.0Å range:
|
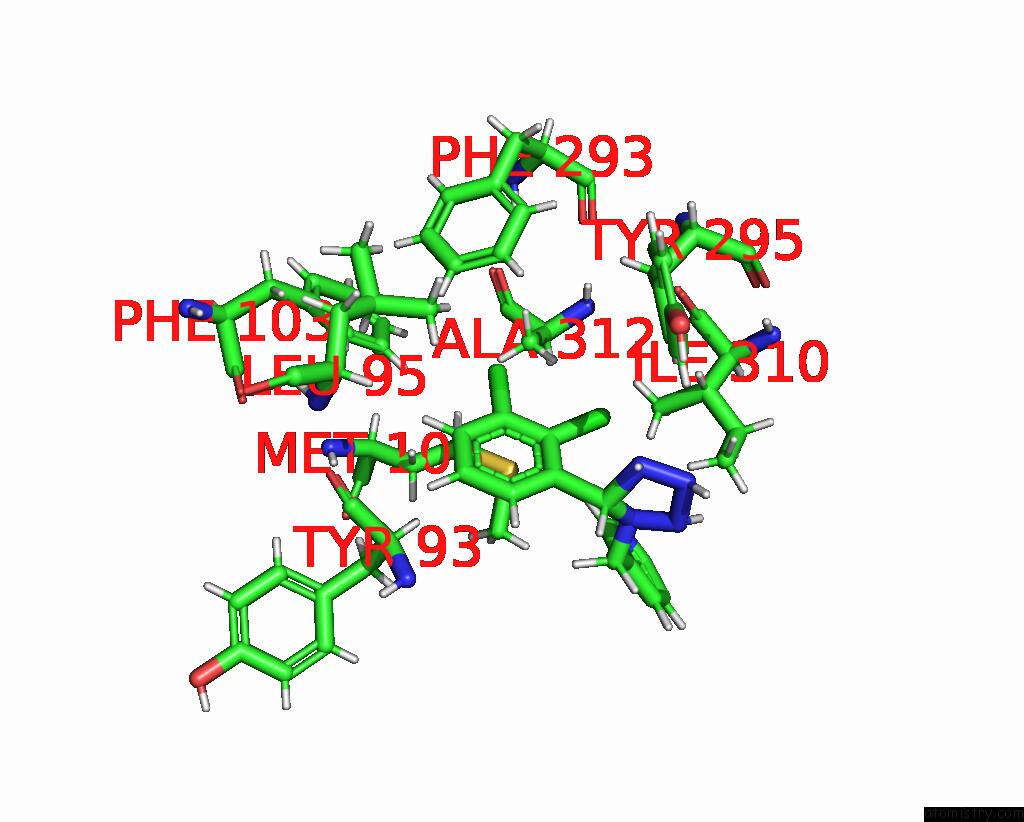
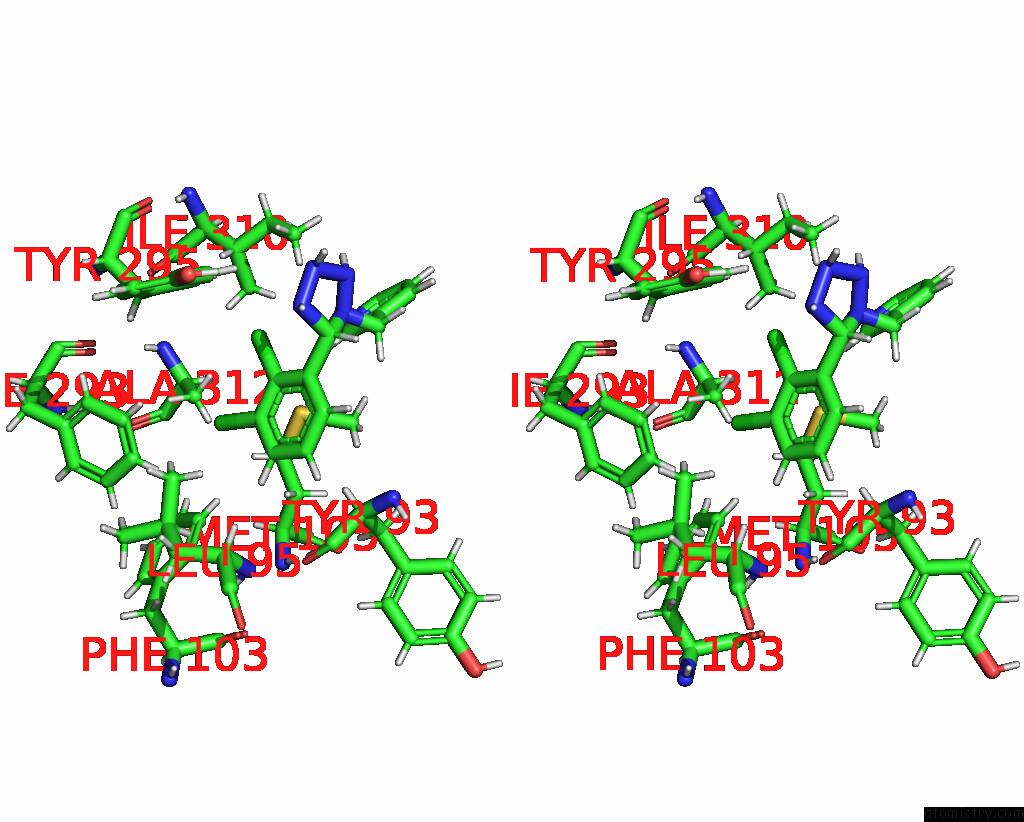
Chlorine binding site 3 out of 6 in 8tr6
Go back to
Chlorine binding site 3 out
of 6 in the Cryo-Em Structure of the Rat P2X7 Receptor in Complex with the Allosteric Antagonist A438079
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Cryo-Em Structure of the Rat P2X7 Receptor in Complex with the Allosteric Antagonist A438079 within 5.0Å range:
|
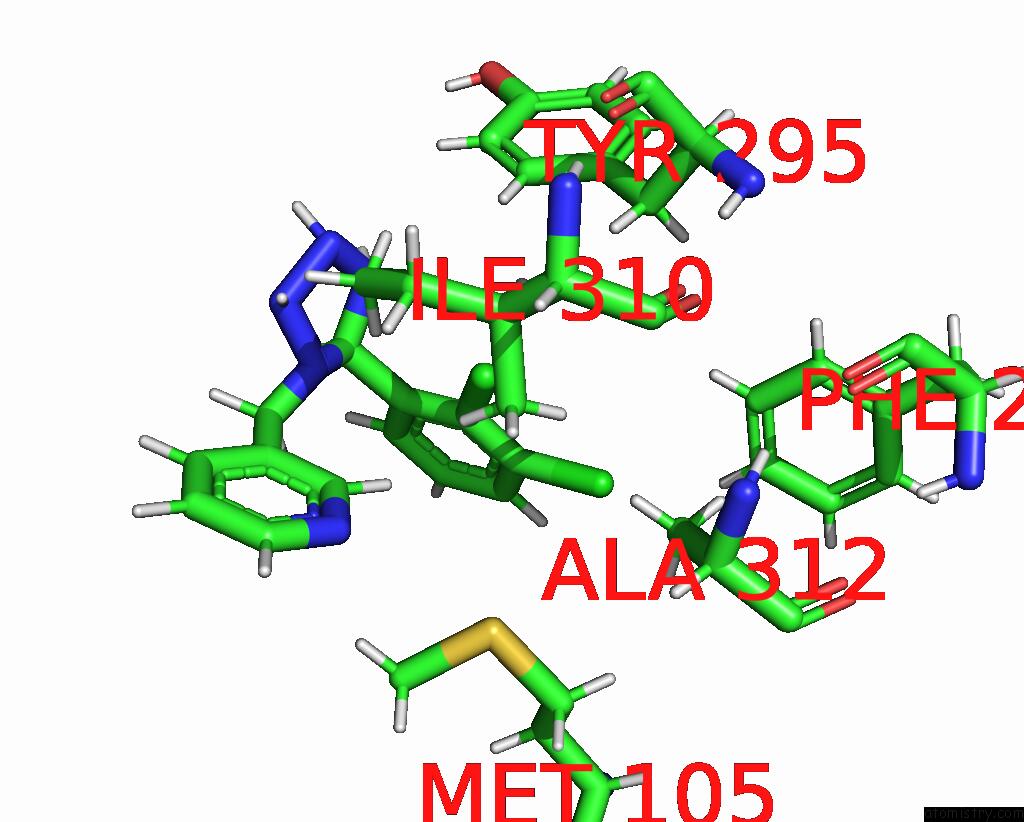
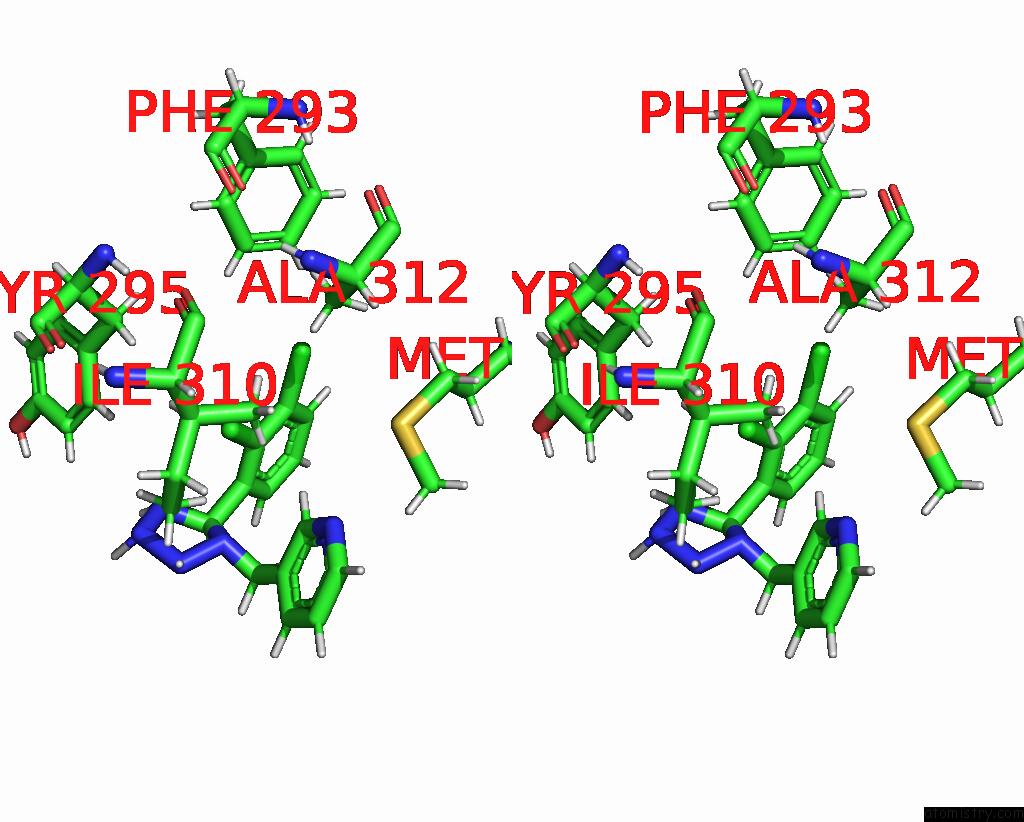
Chlorine binding site 4 out of 6 in 8tr6
Go back to
Chlorine binding site 4 out
of 6 in the Cryo-Em Structure of the Rat P2X7 Receptor in Complex with the Allosteric Antagonist A438079
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Cryo-Em Structure of the Rat P2X7 Receptor in Complex with the Allosteric Antagonist A438079 within 5.0Å range:
|
Chlorine binding site 5 out of 6 in 8tr6
Go back to
Chlorine binding site 5 out
of 6 in the Cryo-Em Structure of the Rat P2X7 Receptor in Complex with the Allosteric Antagonist A438079
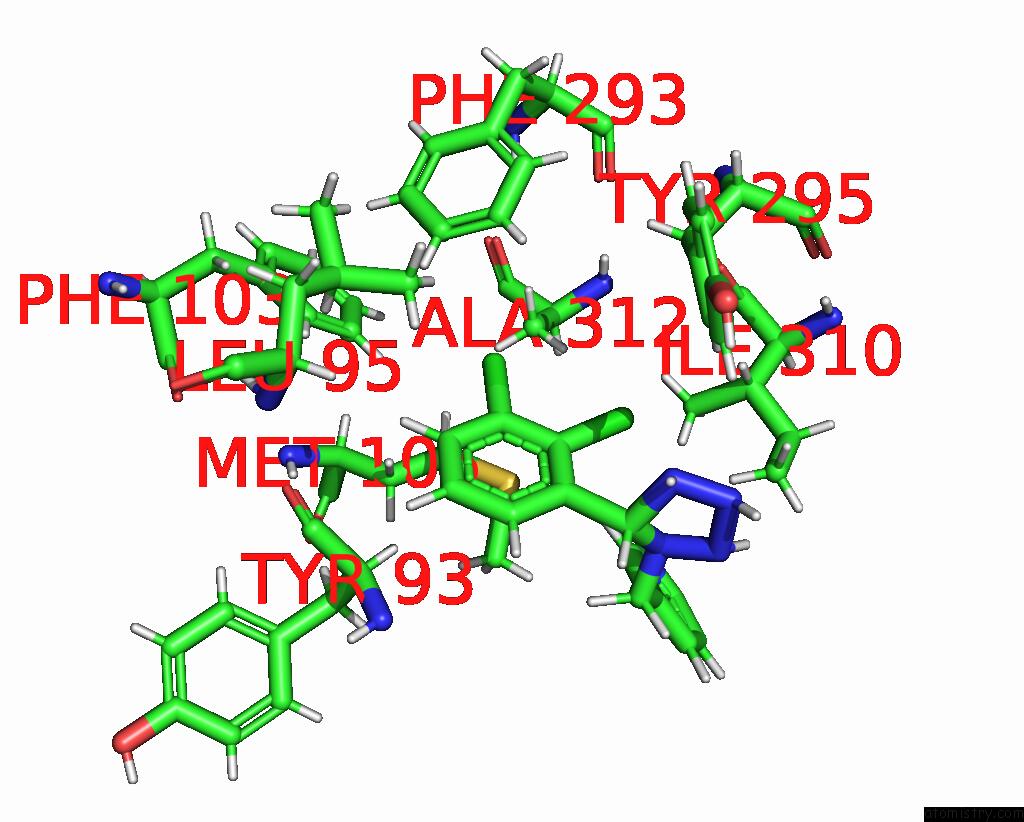
Mono view
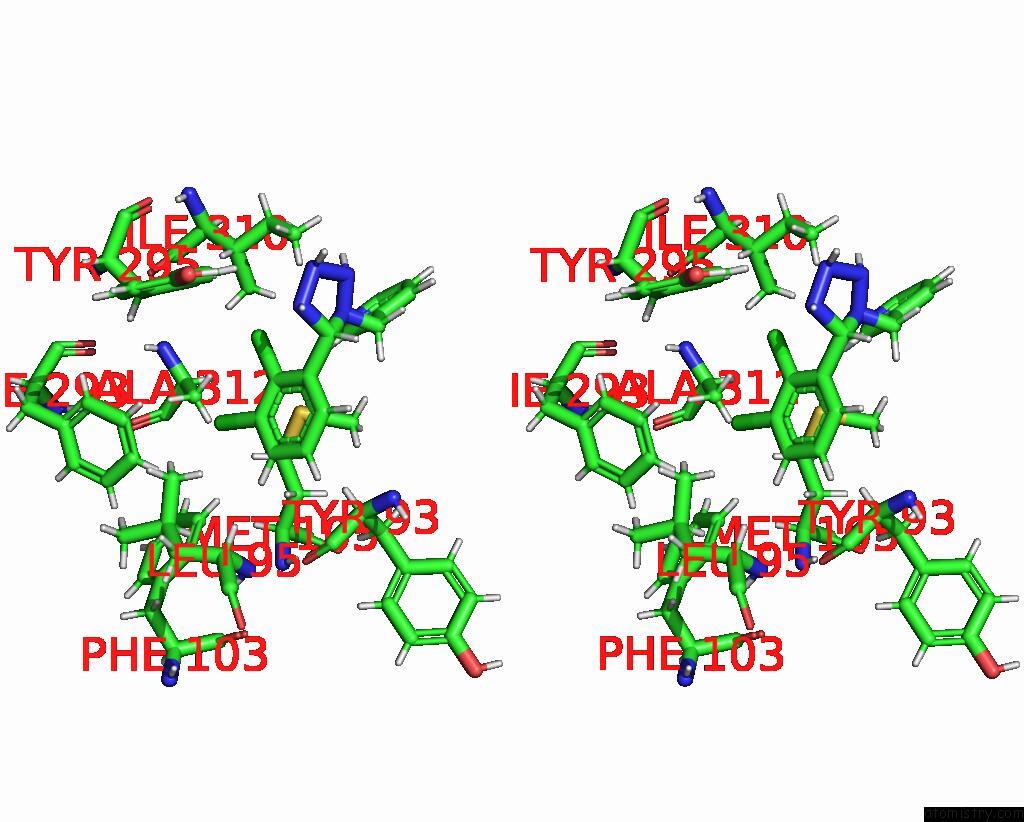
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Cryo-Em Structure of the Rat P2X7 Receptor in Complex with the Allosteric Antagonist A438079 within 5.0Å range:
|
Chlorine binding site 6 out of 6 in 8tr6
Go back to
Chlorine binding site 6 out
of 6 in the Cryo-Em Structure of the Rat P2X7 Receptor in Complex with the Allosteric Antagonist A438079
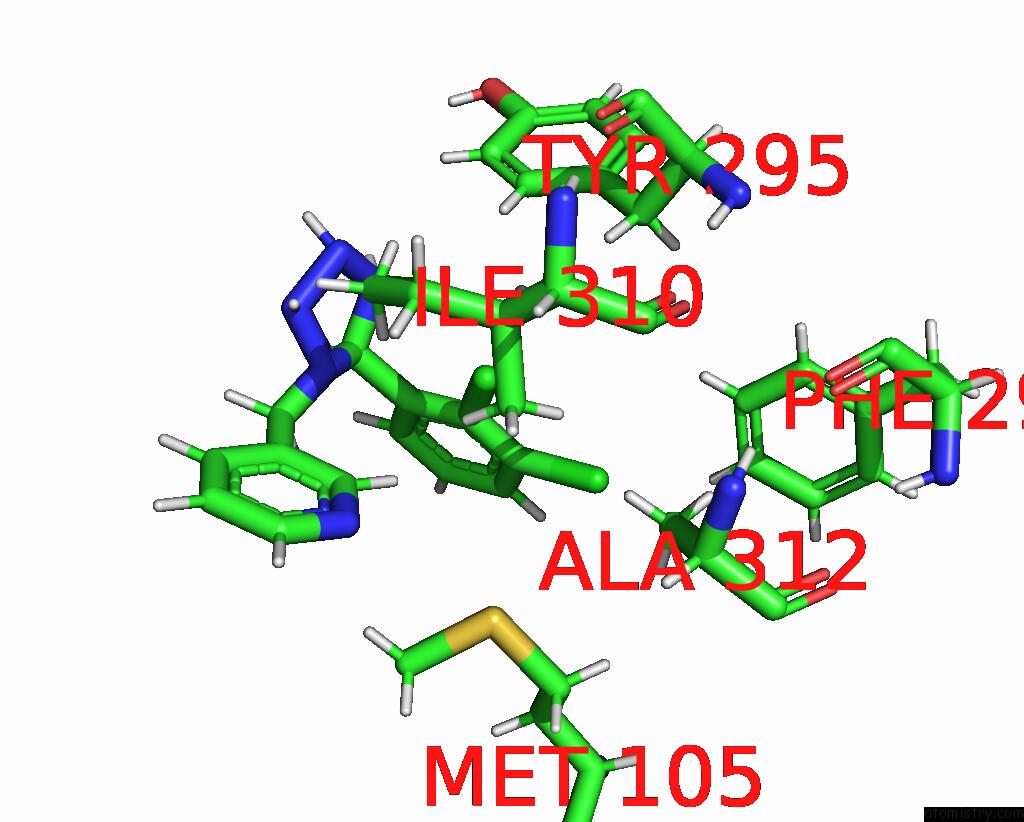
Mono view
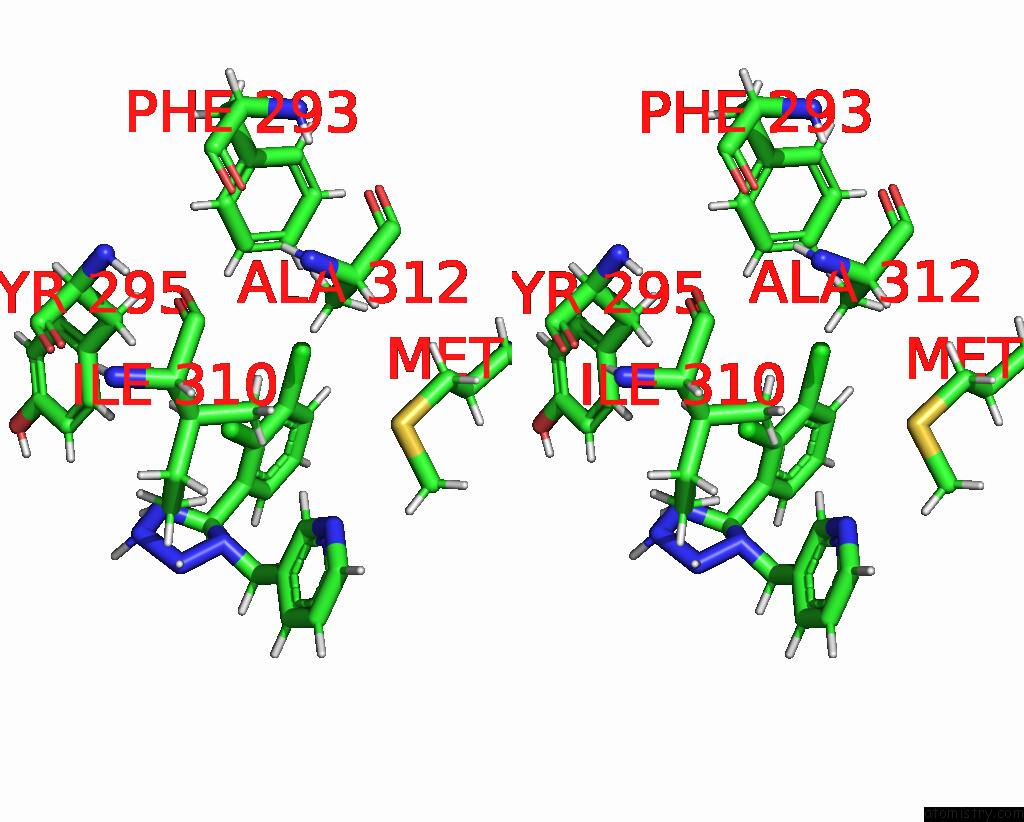
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Cryo-Em Structure of the Rat P2X7 Receptor in Complex with the Allosteric Antagonist A438079 within 5.0Å range:
|
Reference:
A.C.Oken,
I.A.Ditter,
N.E.Lisi,
I.Krishnamurthy,
M.H.Godsey,
S.E.Mansoor.
P2X 7 Receptors Exhibit at Least Three Modes of Allosteric Antagonism. Sci Adv V. 10 O5084 2024.
ISSN: ESSN 2375-2548
PubMed: 39365862
DOI: 10.1126/SCIADV.ADO5084
Page generated: Sun Jul 13 14:31:08 2025
ISSN: ESSN 2375-2548
PubMed: 39365862
DOI: 10.1126/SCIADV.ADO5084
Last articles
F in 4IBIF in 4IAH
F in 4IAE
F in 4I9H
F in 4I9N
F in 4I9O
F in 4IA9
F in 4I89
F in 4I7S
F in 4I87