Chlorine »
PDB 8udm-8uob »
8uhh »
Chlorine in PDB 8uhh: Anti-Phosphohistidine Fab HSC44.Ck.Elbow.20.N32F
Protein crystallography data
The structure of Anti-Phosphohistidine Fab HSC44.Ck.Elbow.20.N32F, PDB code: 8uhh
was solved by
R.Kalagiri,
R.L.Stanfield,
T.Hunter,
I.A.Wilson,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 46.27 / 1.84 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 48.574, 72.072, 152.189, 90, 90, 90 |
| R / Rfree (%) | 20.4 / 23.6 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Anti-Phosphohistidine Fab HSC44.Ck.Elbow.20.N32F
(pdb code 8uhh). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Anti-Phosphohistidine Fab HSC44.Ck.Elbow.20.N32F, PDB code: 8uhh:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Anti-Phosphohistidine Fab HSC44.Ck.Elbow.20.N32F, PDB code: 8uhh:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 8uhh
Go back to
Chlorine binding site 1 out
of 2 in the Anti-Phosphohistidine Fab HSC44.Ck.Elbow.20.N32F
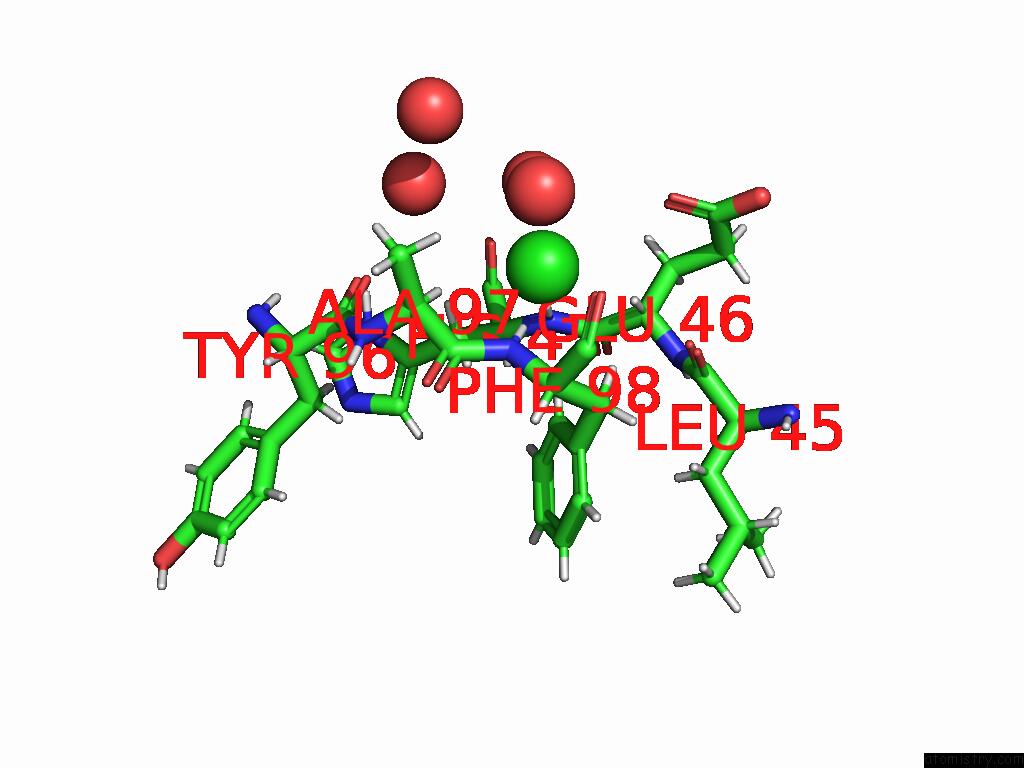
Mono view
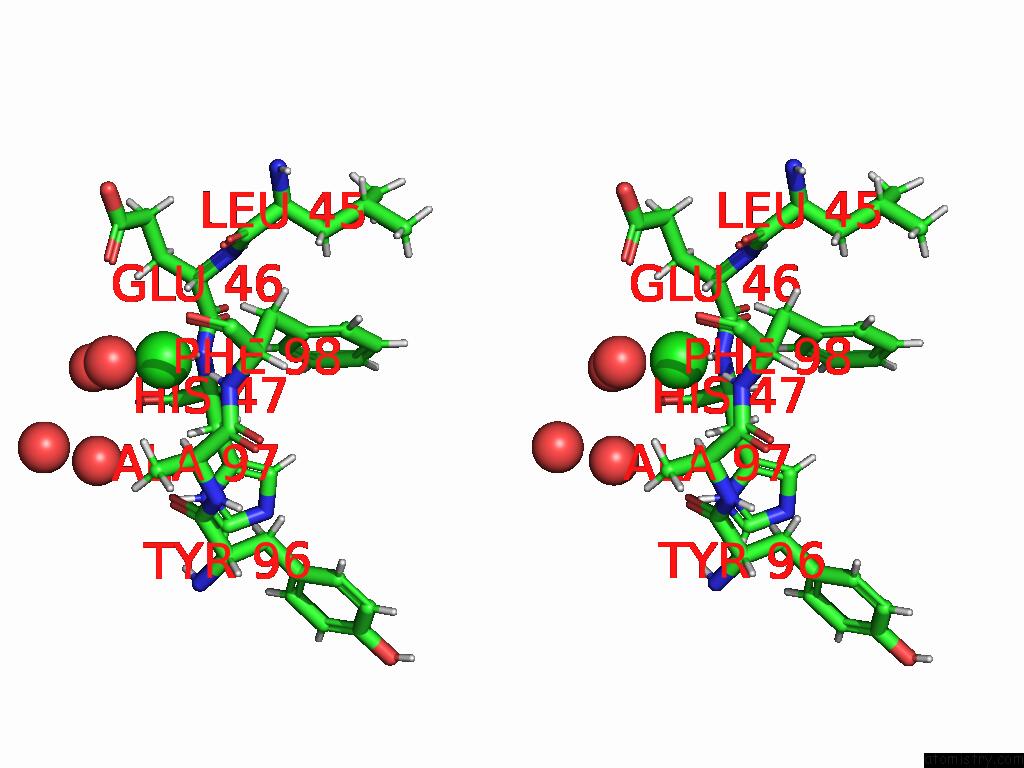
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Anti-Phosphohistidine Fab HSC44.Ck.Elbow.20.N32F within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 8uhh
Go back to
Chlorine binding site 2 out
of 2 in the Anti-Phosphohistidine Fab HSC44.Ck.Elbow.20.N32F
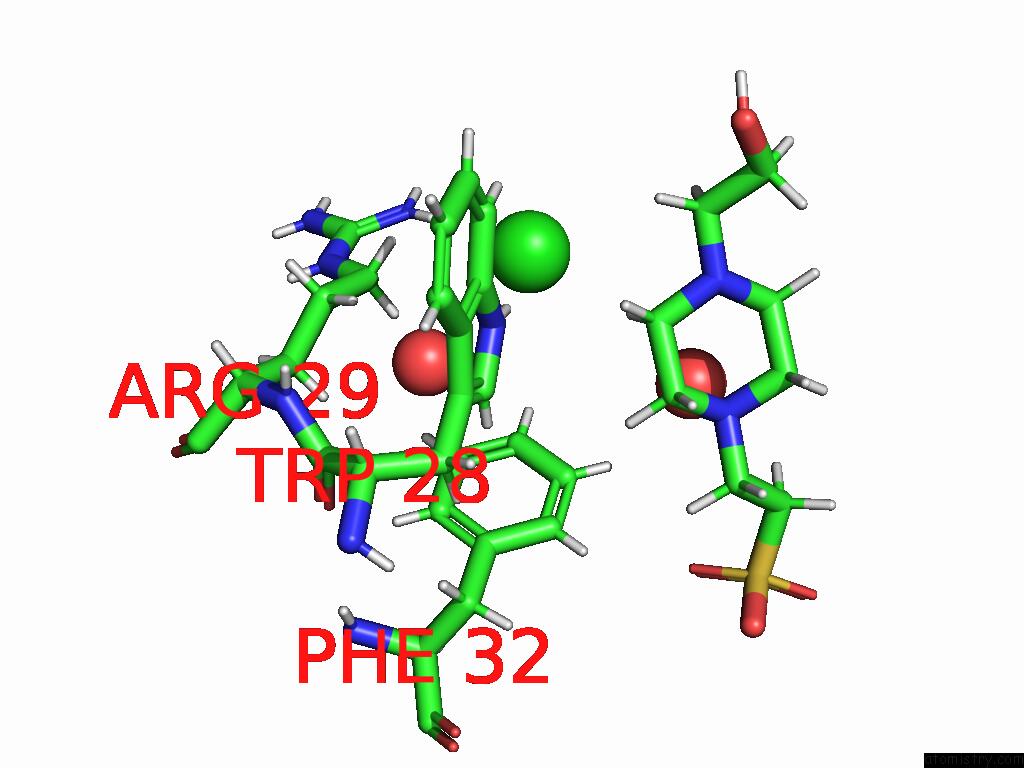
Mono view
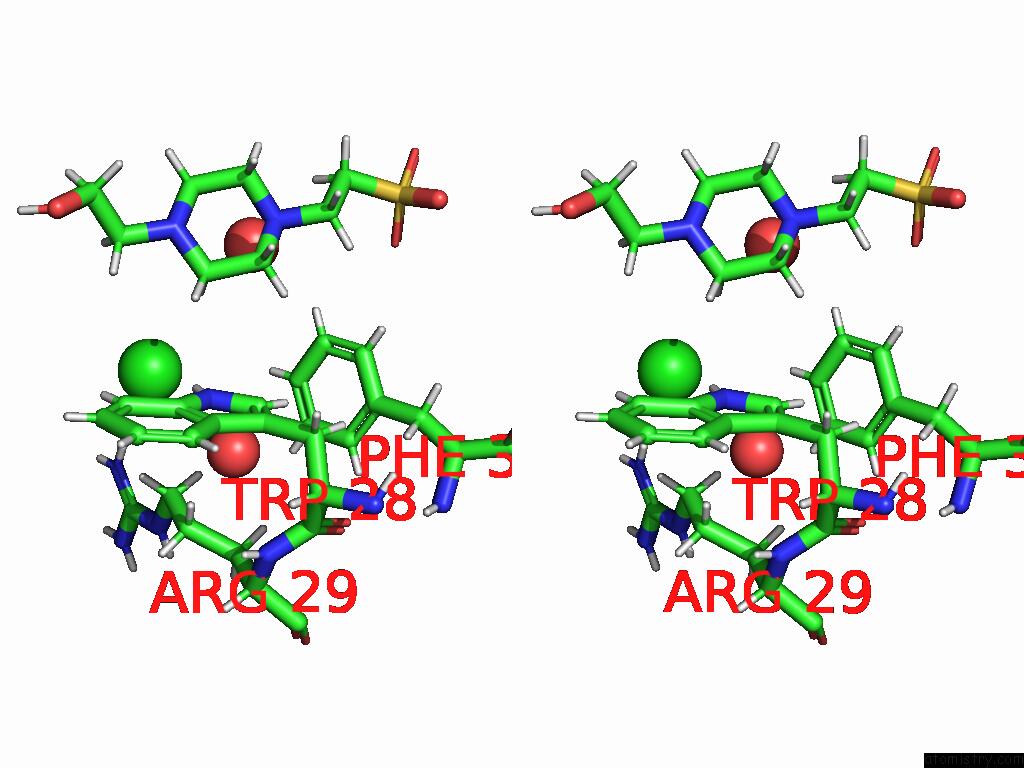
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Anti-Phosphohistidine Fab HSC44.Ck.Elbow.20.N32F within 5.0Å range:
|
Reference:
G.D.Martyn,
R.Kalagiri,
G.Veggiani,
R.L.Stanfield,
I.Choudhuri,
M.Sala,
J.Meisenhelder,
C.Chen,
A.Biswas,
R.M.Levy,
D.Lyumkis,
I.A.Wilson,
T.Hunter,
S.S.Sidhu.
Using Phage Display For Rational Engineering of A Higher Affinity Humanized 3' Phosphohistidine-Specific Antibody. Biorxiv 2024.
ISSN: ISSN 2692-8205
PubMed: 39574610
DOI: 10.1101/2024.11.04.621849
Page generated: Sun Jul 13 14:56:42 2025
ISSN: ISSN 2692-8205
PubMed: 39574610
DOI: 10.1101/2024.11.04.621849
Last articles
Co in 8ASMCo in 7ZZL
Co in 7Z0V
Co in 7Y8P
Co in 7YVX
Co in 7YGP
Co in 7XRN
Co in 7XVH
Co in 7XRM
Co in 7XRL