Chlorine »
PDB 8ux6-8vev »
8v80 »
Chlorine in PDB 8v80: ALPHA7-Nicotinic Acetylcholine Receptor Bound to Epibatidine and (-)- Tqs
Other elements in 8v80:
The structure of ALPHA7-Nicotinic Acetylcholine Receptor Bound to Epibatidine and (-)- Tqs also contains other interesting chemical elements:
| Calcium | (Ca) | 5 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the ALPHA7-Nicotinic Acetylcholine Receptor Bound to Epibatidine and (-)- Tqs
(pdb code 8v80). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 5 binding sites of Chlorine where determined in the ALPHA7-Nicotinic Acetylcholine Receptor Bound to Epibatidine and (-)- Tqs, PDB code: 8v80:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Chlorine where determined in the ALPHA7-Nicotinic Acetylcholine Receptor Bound to Epibatidine and (-)- Tqs, PDB code: 8v80:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5;
Chlorine binding site 1 out of 5 in 8v80
Go back to
Chlorine binding site 1 out
of 5 in the ALPHA7-Nicotinic Acetylcholine Receptor Bound to Epibatidine and (-)- Tqs
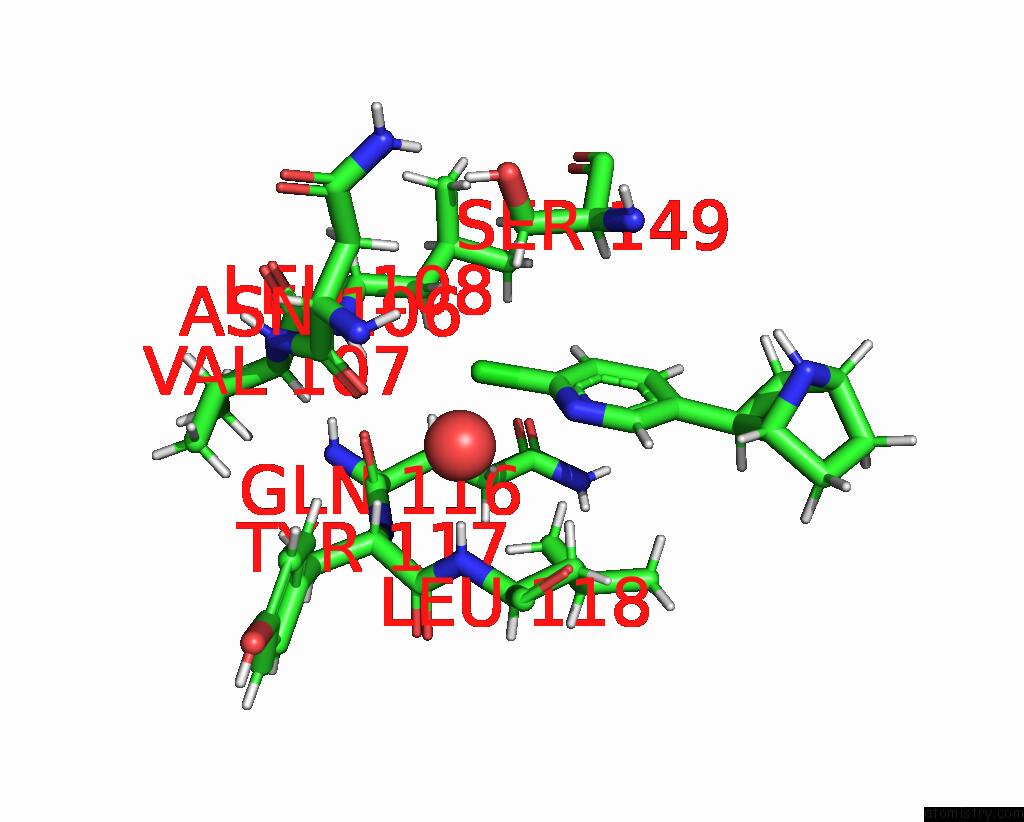
Mono view
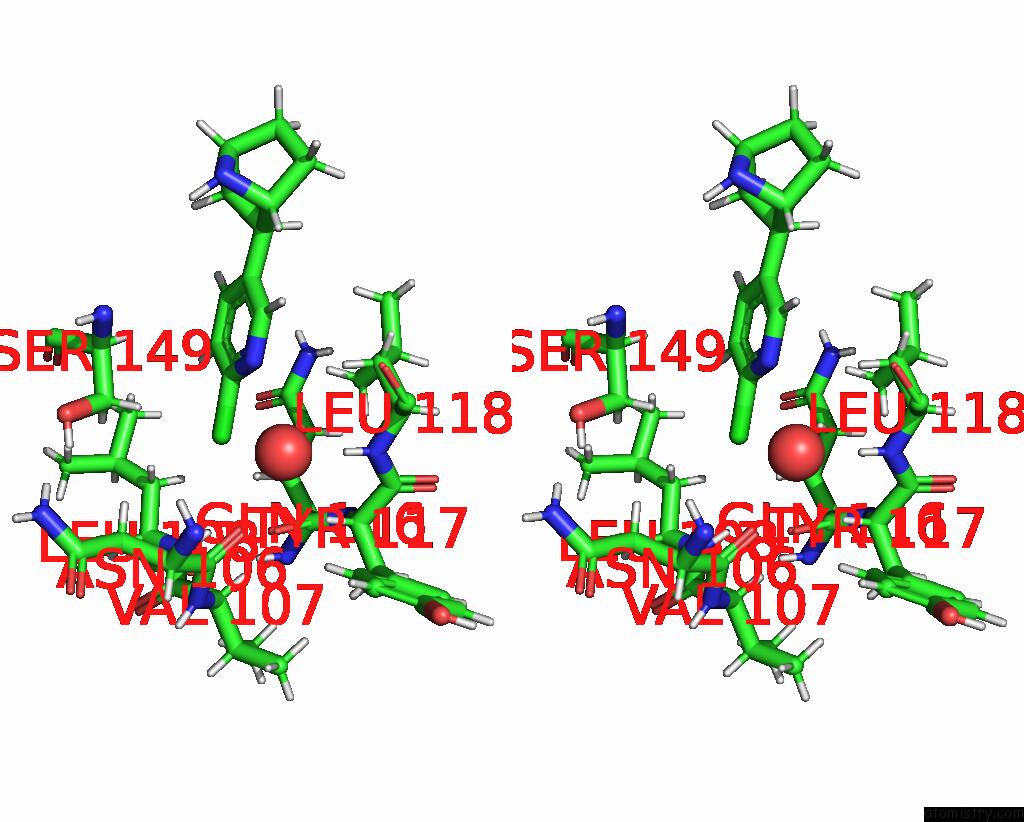
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of ALPHA7-Nicotinic Acetylcholine Receptor Bound to Epibatidine and (-)- Tqs within 5.0Å range:
|
Chlorine binding site 2 out of 5 in 8v80
Go back to
Chlorine binding site 2 out
of 5 in the ALPHA7-Nicotinic Acetylcholine Receptor Bound to Epibatidine and (-)- Tqs
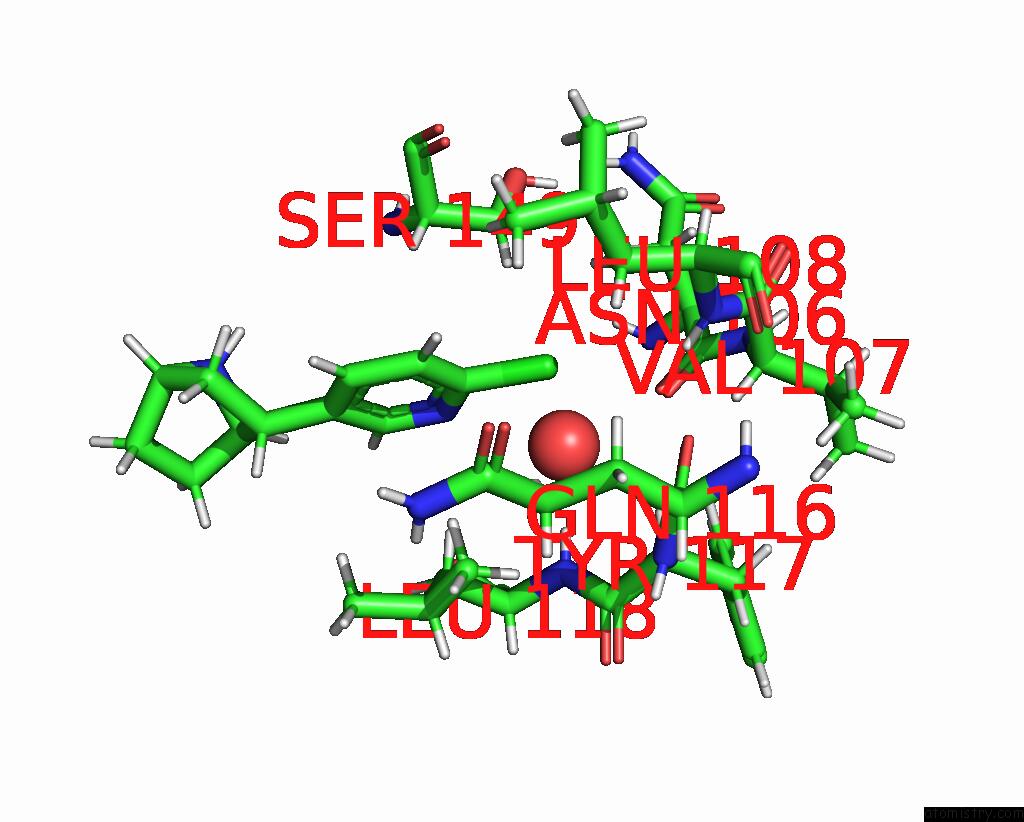
Mono view
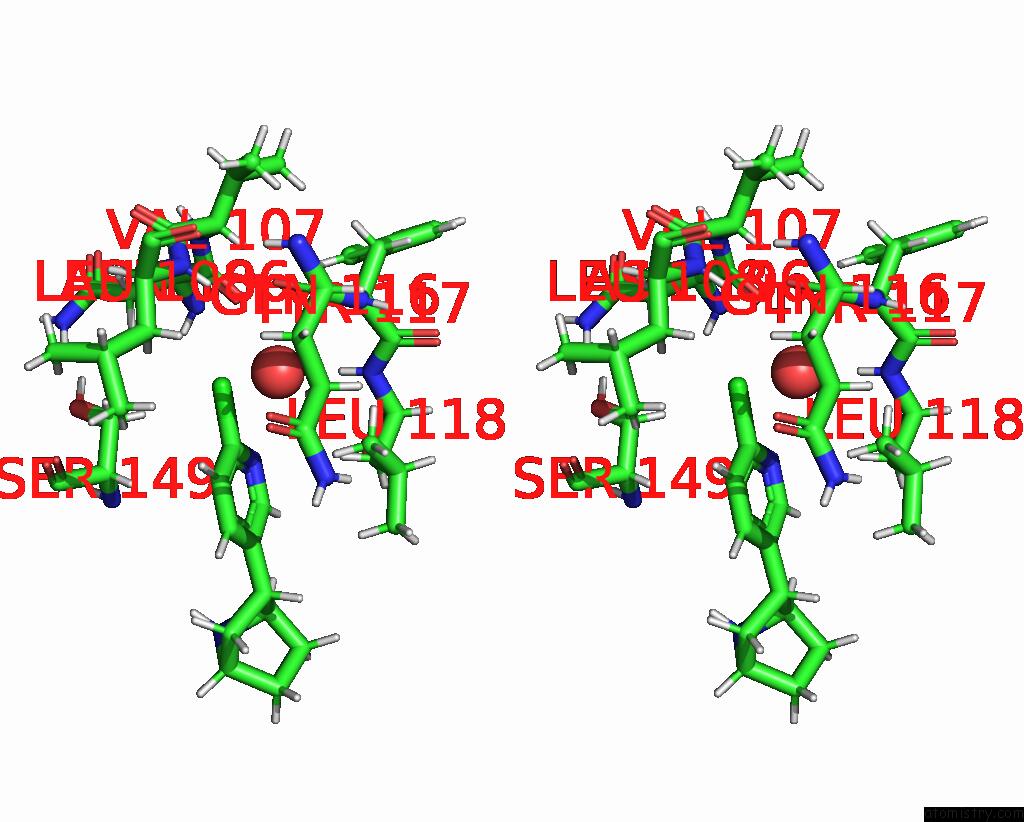
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of ALPHA7-Nicotinic Acetylcholine Receptor Bound to Epibatidine and (-)- Tqs within 5.0Å range:
|
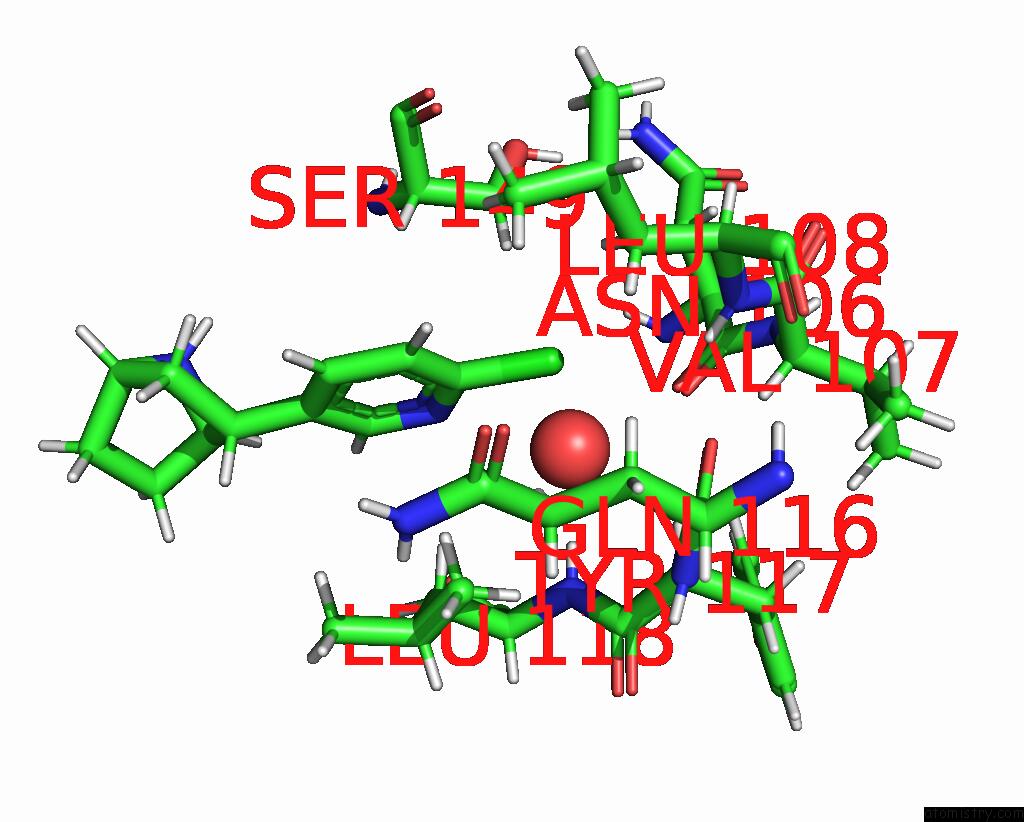
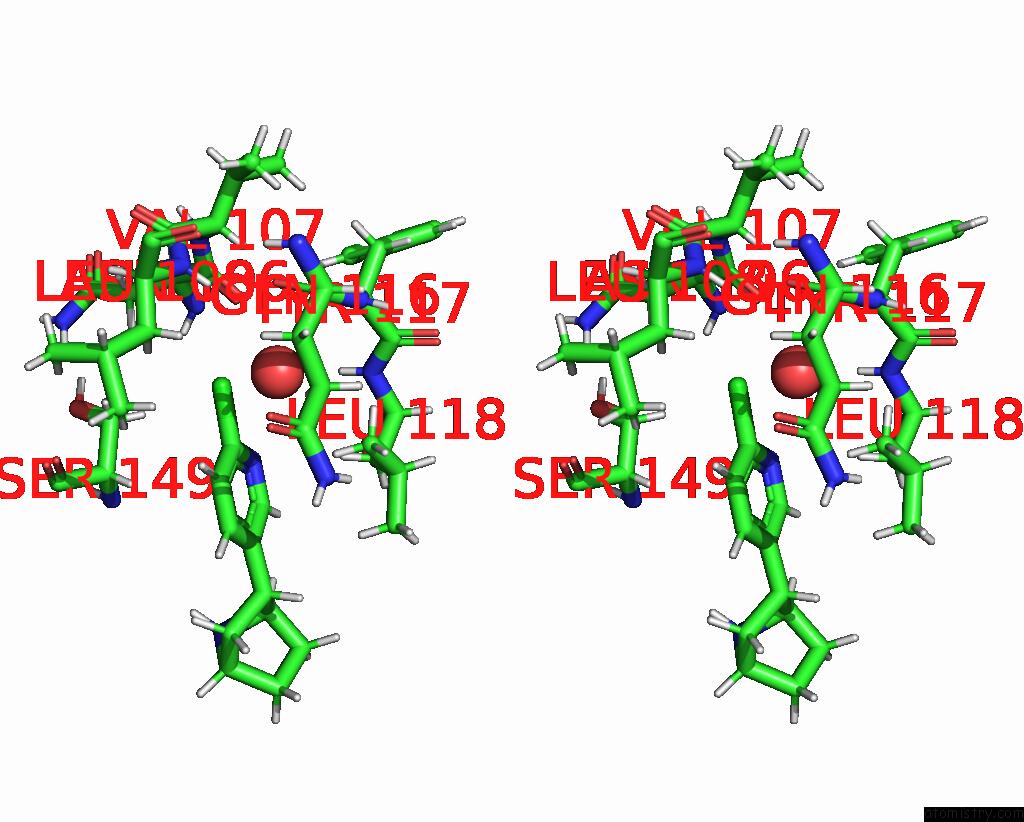
Chlorine binding site 3 out of 5 in 8v80
Go back to
Chlorine binding site 3 out
of 5 in the ALPHA7-Nicotinic Acetylcholine Receptor Bound to Epibatidine and (-)- Tqs
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of ALPHA7-Nicotinic Acetylcholine Receptor Bound to Epibatidine and (-)- Tqs within 5.0Å range:
|
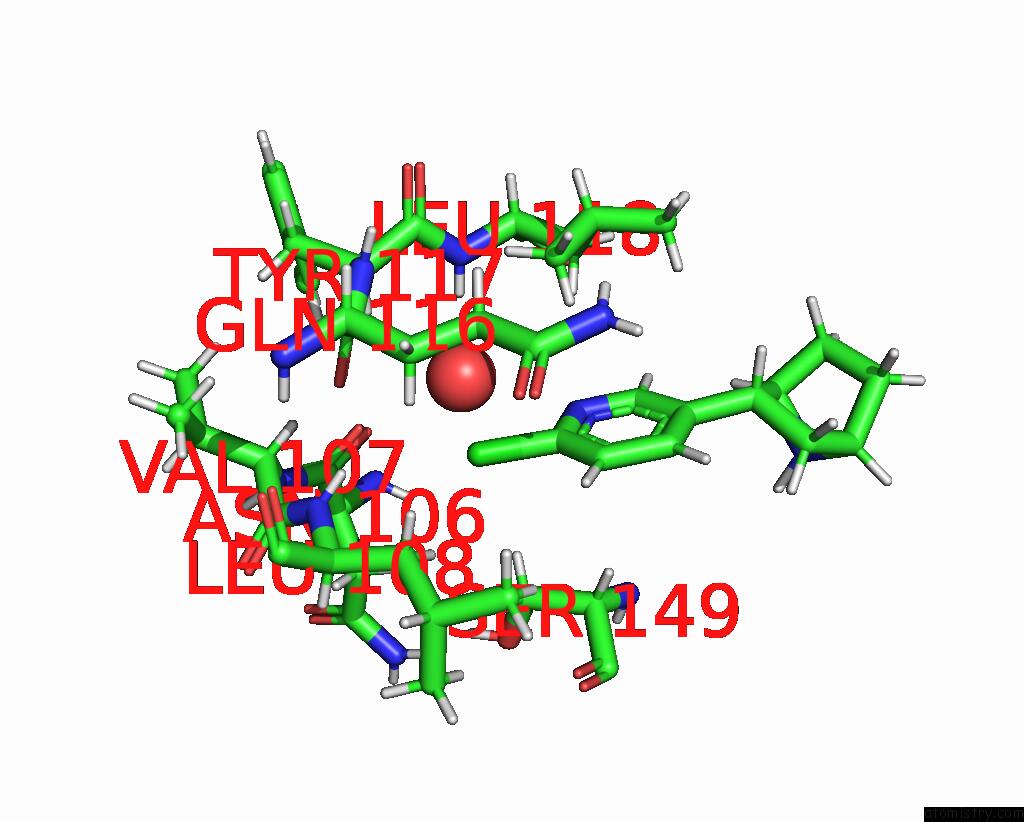
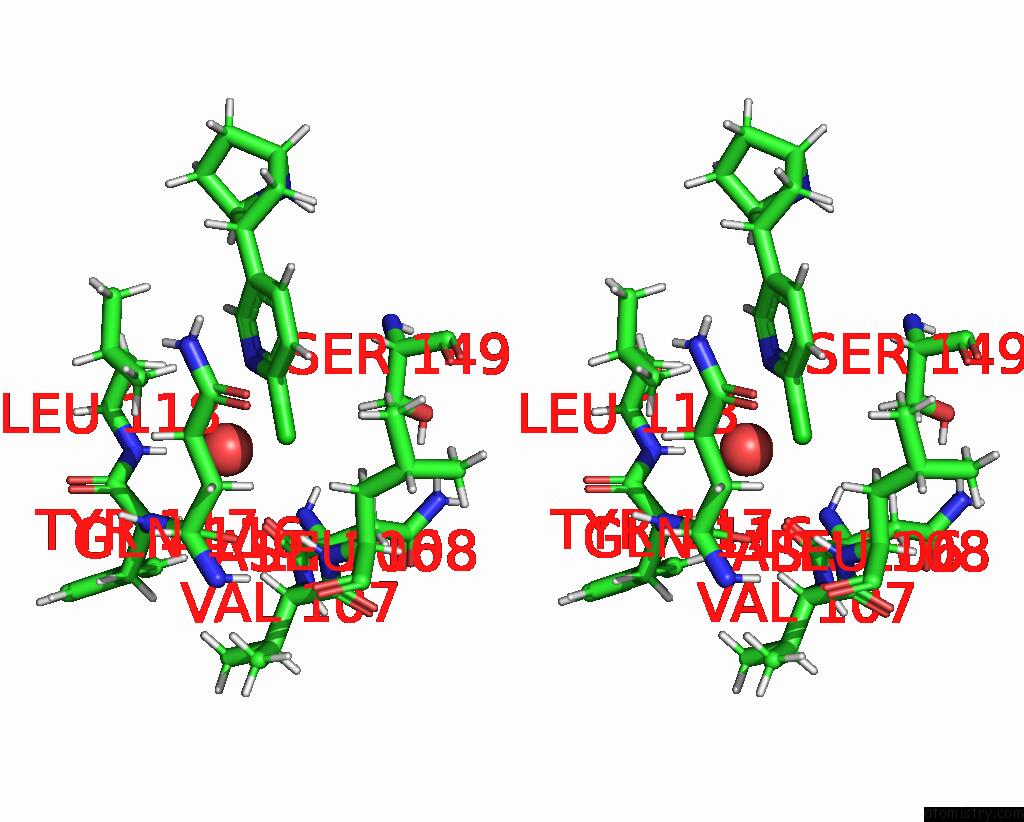
Chlorine binding site 4 out of 5 in 8v80
Go back to
Chlorine binding site 4 out
of 5 in the ALPHA7-Nicotinic Acetylcholine Receptor Bound to Epibatidine and (-)- Tqs
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of ALPHA7-Nicotinic Acetylcholine Receptor Bound to Epibatidine and (-)- Tqs within 5.0Å range:
|
Chlorine binding site 5 out of 5 in 8v80
Go back to
Chlorine binding site 5 out
of 5 in the ALPHA7-Nicotinic Acetylcholine Receptor Bound to Epibatidine and (-)- Tqs
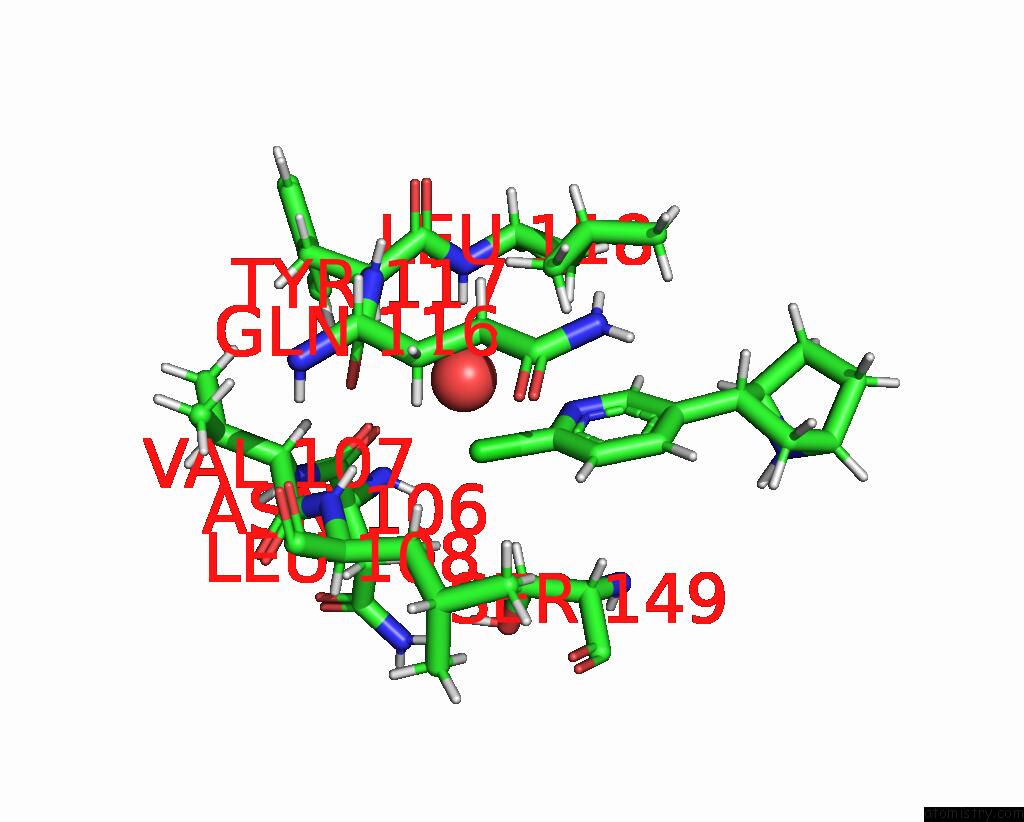
Mono view
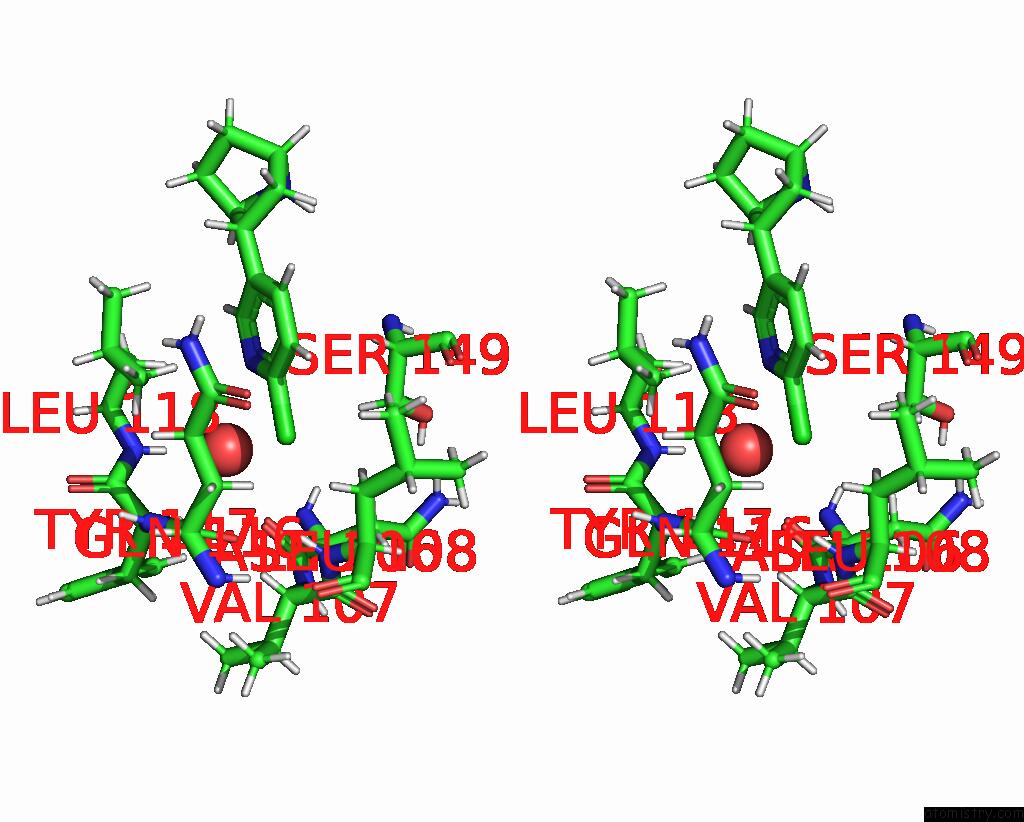
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of ALPHA7-Nicotinic Acetylcholine Receptor Bound to Epibatidine and (-)- Tqs within 5.0Å range:
|
Reference:
S.M.Burke,
M.Avstrikova,
C.M.Noviello,
N.Mukhtasimova,
J.P.Changeux,
G.A.Thakur,
S.M.Sine,
M.Cecchini,
R.E.Hibbs.
Structural Mechanisms of ALPHA7 Nicotinic Receptor Allosteric Modulation and Activation To Be Published.
Page generated: Sun Jul 13 15:11:29 2025
Last articles
F in 7PRXF in 7PPH
F in 7PRM
F in 7PQV
F in 7PQS
F in 7PMM
F in 7POL
F in 7POR
F in 7PNS
F in 7P81