Chlorine »
PDB 9cr5-9dpx »
9db5 »
Chlorine in PDB 9db5: A Darpin Fused to the 1TEL Crystallization Chaperone Via A Proline- Alanine Linker
Protein crystallography data
The structure of A Darpin Fused to the 1TEL Crystallization Chaperone Via A Proline- Alanine Linker, PDB code: 9db5
was solved by
M.J.Pedroza Romo,
J.C.Averett,
A.Keliiliki,
E.W.Wilson,
C.Smith,
D.Hansen,
B.Averett,
J.Gonzalez,
E.Noakes,
R.Nickles,
T.Doukov,
J.D.Moody,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.87 / 1.57 |
| Space group | P 65 |
| Cell size a, b, c (Å), α, β, γ (°) | 97.73, 97.73, 45.816, 90, 90, 120 |
| R / Rfree (%) | 16.7 / 19.6 |
Other elements in 9db5:
The structure of A Darpin Fused to the 1TEL Crystallization Chaperone Via A Proline- Alanine Linker also contains other interesting chemical elements:
| Sodium | (Na) | 8 atoms |
Chlorine Binding Sites:
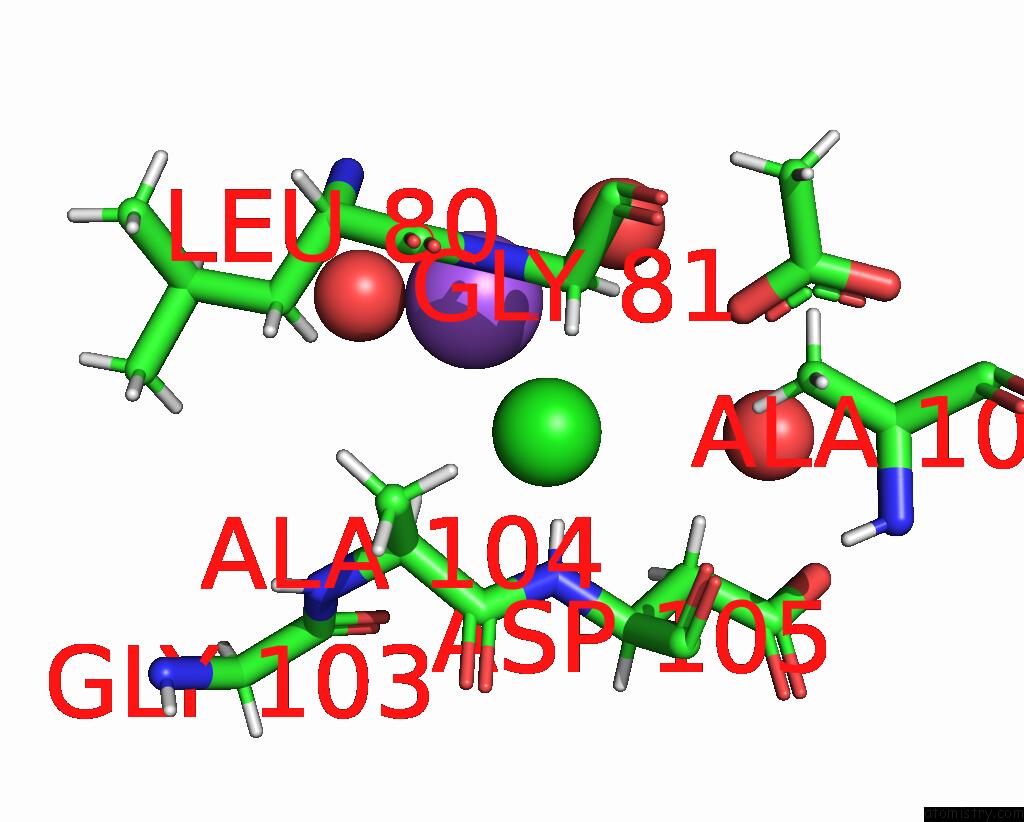
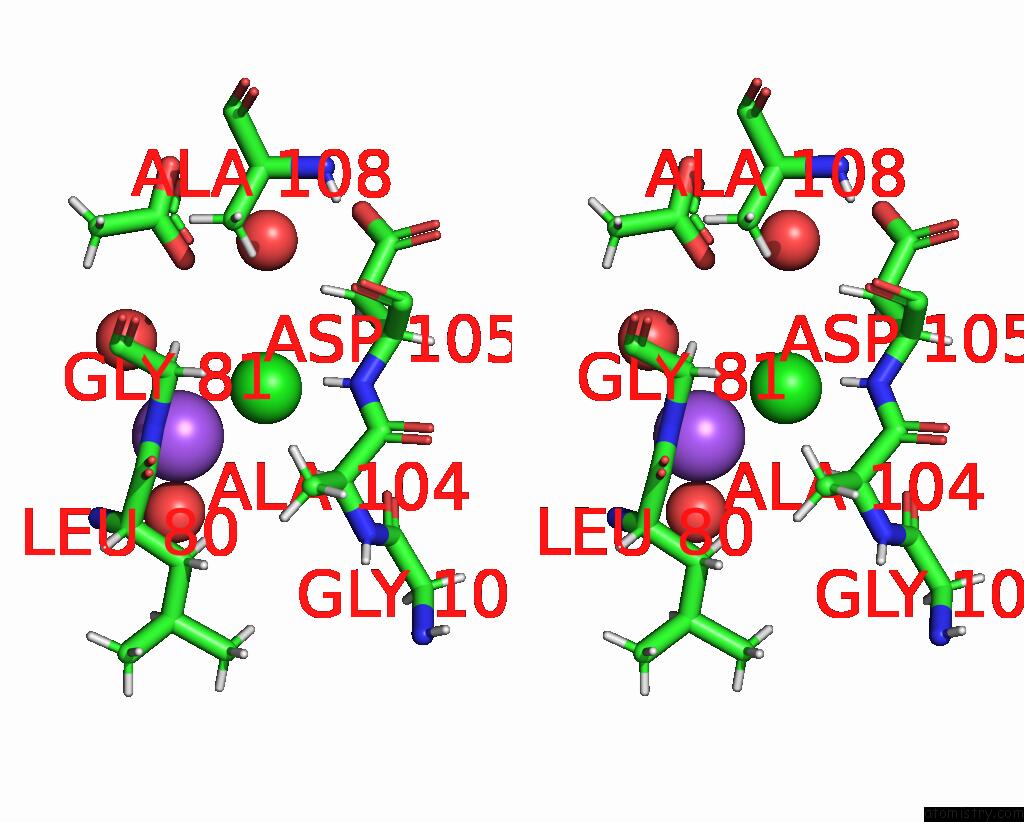
The binding sites of Chlorine atom in the A Darpin Fused to the 1TEL Crystallization Chaperone Via A Proline- Alanine Linker
(pdb code 9db5). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the A Darpin Fused to the 1TEL Crystallization Chaperone Via A Proline- Alanine Linker, PDB code: 9db5:
In total only one binding site of Chlorine was determined in the A Darpin Fused to the 1TEL Crystallization Chaperone Via A Proline- Alanine Linker, PDB code: 9db5:
Chlorine binding site 1 out of 1 in 9db5
Go back to
Chlorine binding site 1 out
of 1 in the A Darpin Fused to the 1TEL Crystallization Chaperone Via A Proline- Alanine Linker
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of A Darpin Fused to the 1TEL Crystallization Chaperone Via A Proline- Alanine Linker within 5.0Å range:
|
Reference:
M.J.Pedroza Romo,
J.C.Averett,
A.Keliiliki,
E.W.Wilson,
C.Smith,
D.Hansen,
B.Averett,
J.Gonzalez,
E.Noakes,
R.Nickles,
T.Doukov,
J.D.Moody.
Optimal Telsam-Target Protein Linker Character Is Target Protein Dependent To Be Published.
Page generated: Sun Jul 13 16:15:55 2025
Last articles
F in 4HLHF in 4HL4
F in 4HIQ
F in 4HHZ
F in 4HHY
F in 4HGT
F in 4HEJ
F in 4HGS
F in 4H52
F in 4H4O