Chlorine »
PDB 9fex-9g0g »
9ff2 »
Chlorine in PDB 9ff2: Cryo-Em Structure of the BETA3 Homomeric Gaba(A) Receptor in Complex with Hsm in the Long-Lived Symmetric Desensitised State (C1)
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Cryo-Em Structure of the BETA3 Homomeric Gaba(A) Receptor in Complex with Hsm in the Long-Lived Symmetric Desensitised State (C1)
(pdb code 9ff2). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 5 binding sites of Chlorine where determined in the Cryo-Em Structure of the BETA3 Homomeric Gaba(A) Receptor in Complex with Hsm in the Long-Lived Symmetric Desensitised State (C1), PDB code: 9ff2:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Chlorine where determined in the Cryo-Em Structure of the BETA3 Homomeric Gaba(A) Receptor in Complex with Hsm in the Long-Lived Symmetric Desensitised State (C1), PDB code: 9ff2:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5;
Chlorine binding site 1 out of 5 in 9ff2
Go back to
Chlorine binding site 1 out
of 5 in the Cryo-Em Structure of the BETA3 Homomeric Gaba(A) Receptor in Complex with Hsm in the Long-Lived Symmetric Desensitised State (C1)
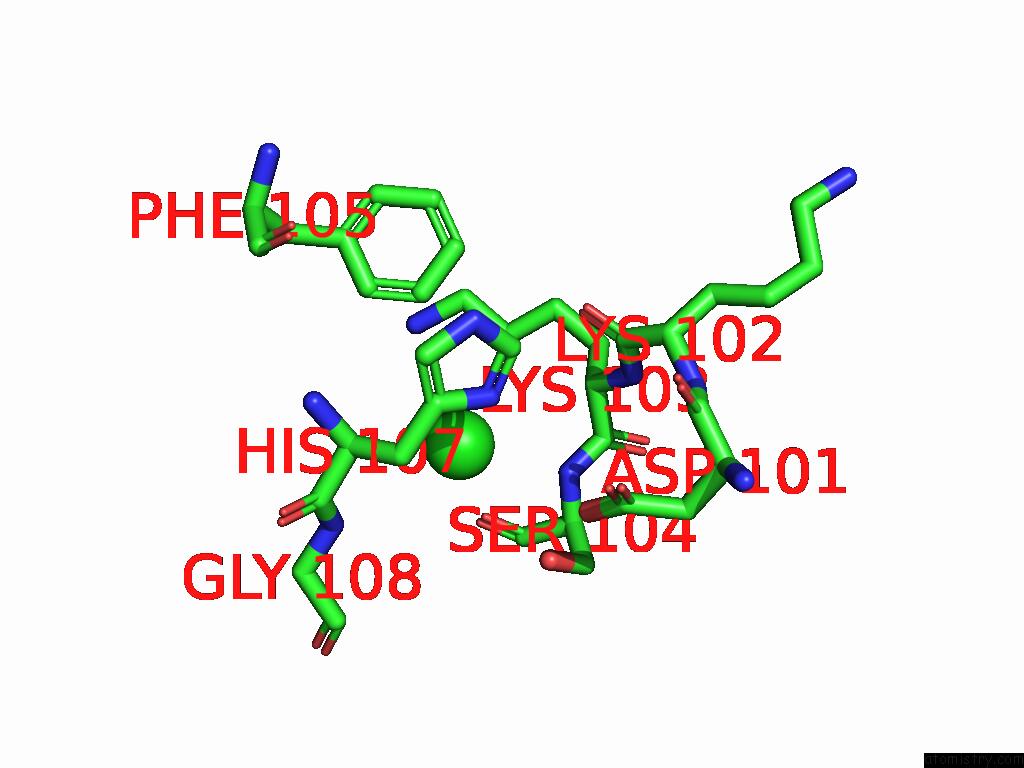
Mono view
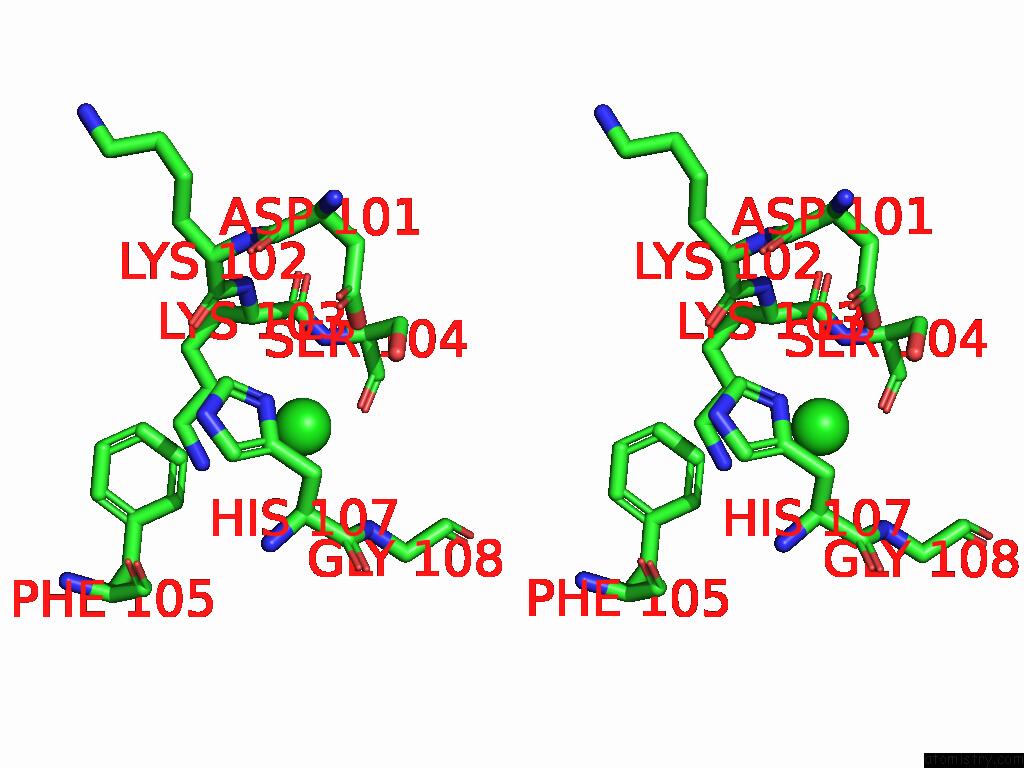
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Cryo-Em Structure of the BETA3 Homomeric Gaba(A) Receptor in Complex with Hsm in the Long-Lived Symmetric Desensitised State (C1) within 5.0Å range:
|
Chlorine binding site 2 out of 5 in 9ff2
Go back to
Chlorine binding site 2 out
of 5 in the Cryo-Em Structure of the BETA3 Homomeric Gaba(A) Receptor in Complex with Hsm in the Long-Lived Symmetric Desensitised State (C1)
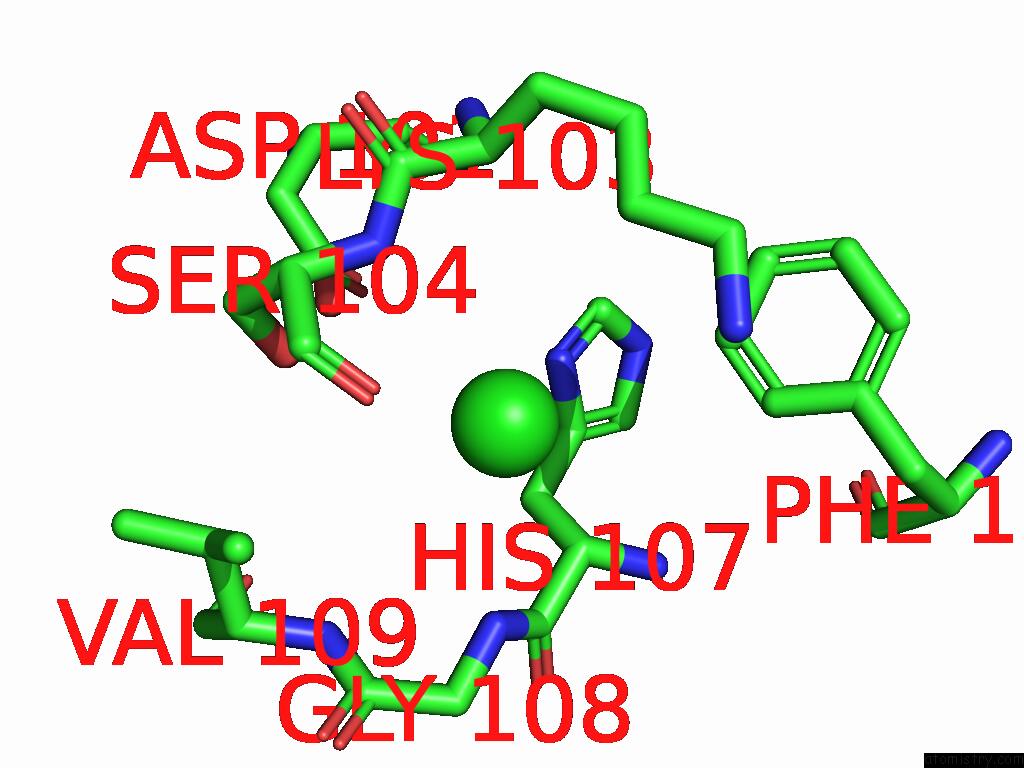
Mono view
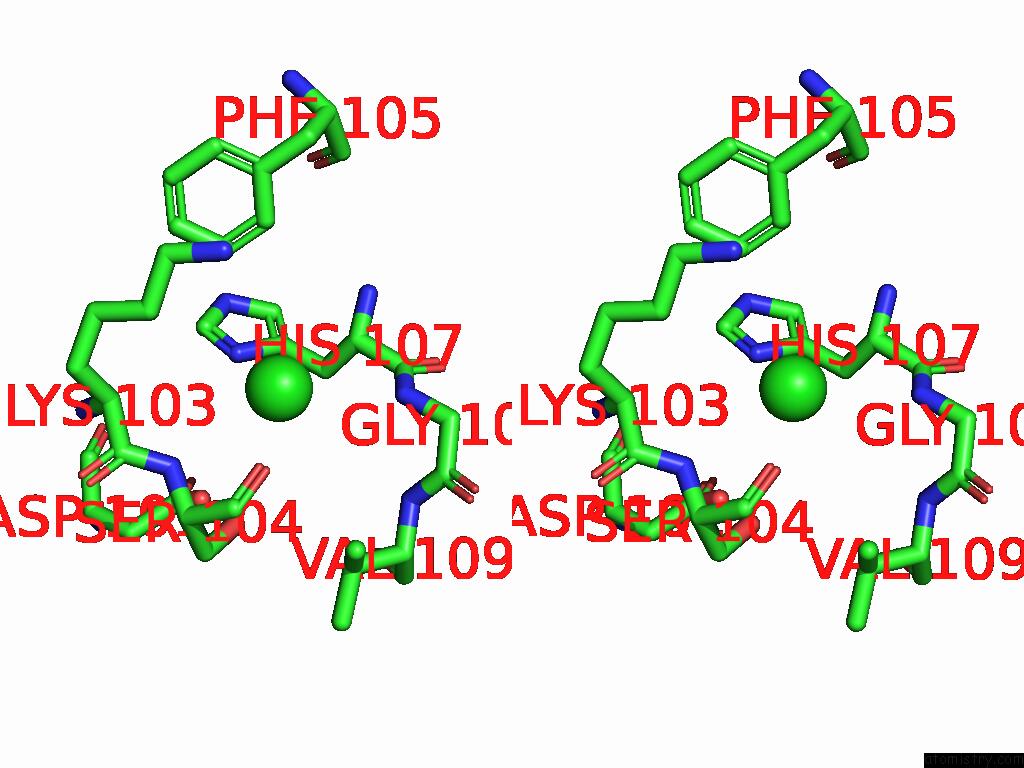
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Cryo-Em Structure of the BETA3 Homomeric Gaba(A) Receptor in Complex with Hsm in the Long-Lived Symmetric Desensitised State (C1) within 5.0Å range:
|
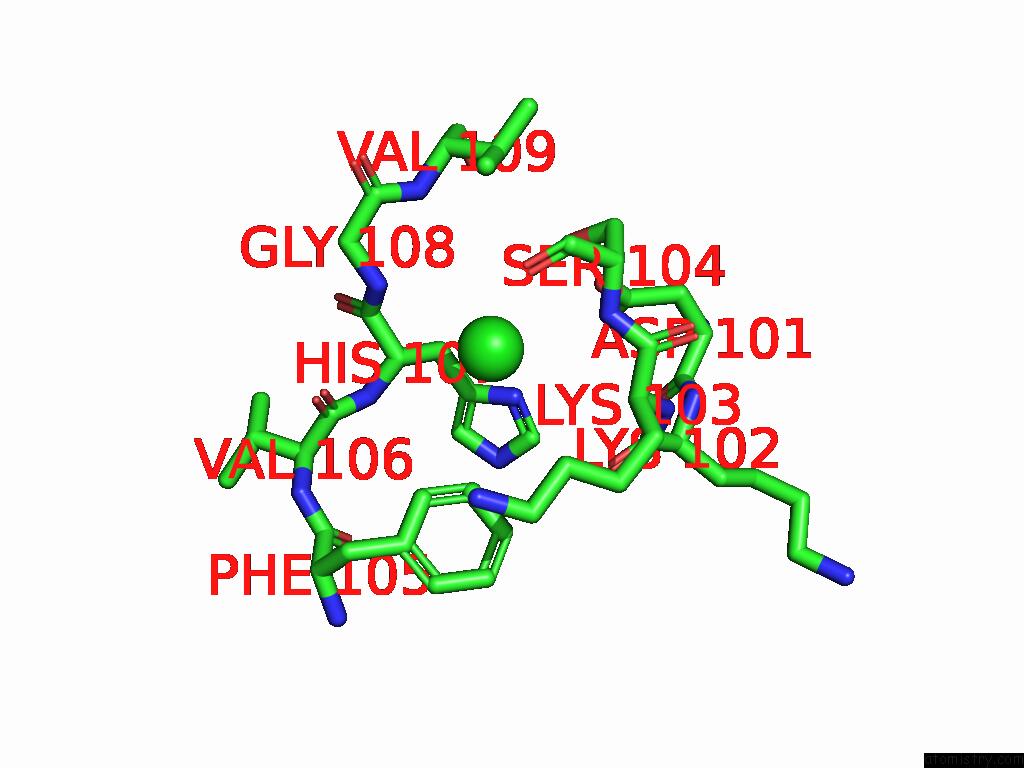
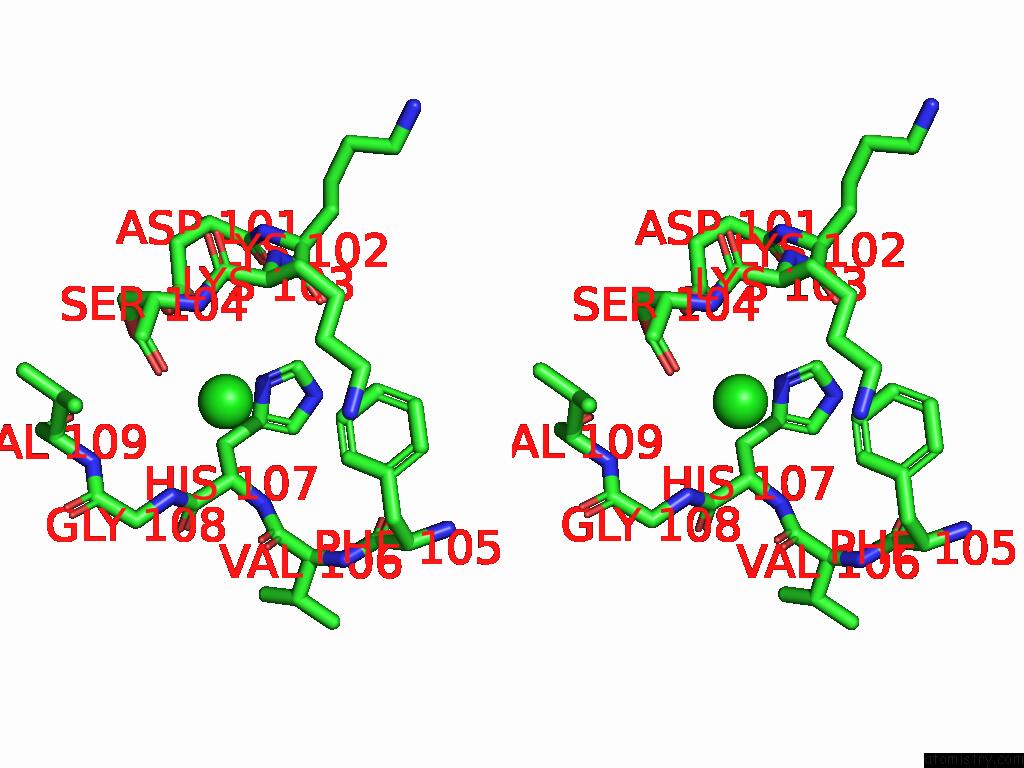
Chlorine binding site 3 out of 5 in 9ff2
Go back to
Chlorine binding site 3 out
of 5 in the Cryo-Em Structure of the BETA3 Homomeric Gaba(A) Receptor in Complex with Hsm in the Long-Lived Symmetric Desensitised State (C1)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Cryo-Em Structure of the BETA3 Homomeric Gaba(A) Receptor in Complex with Hsm in the Long-Lived Symmetric Desensitised State (C1) within 5.0Å range:
|
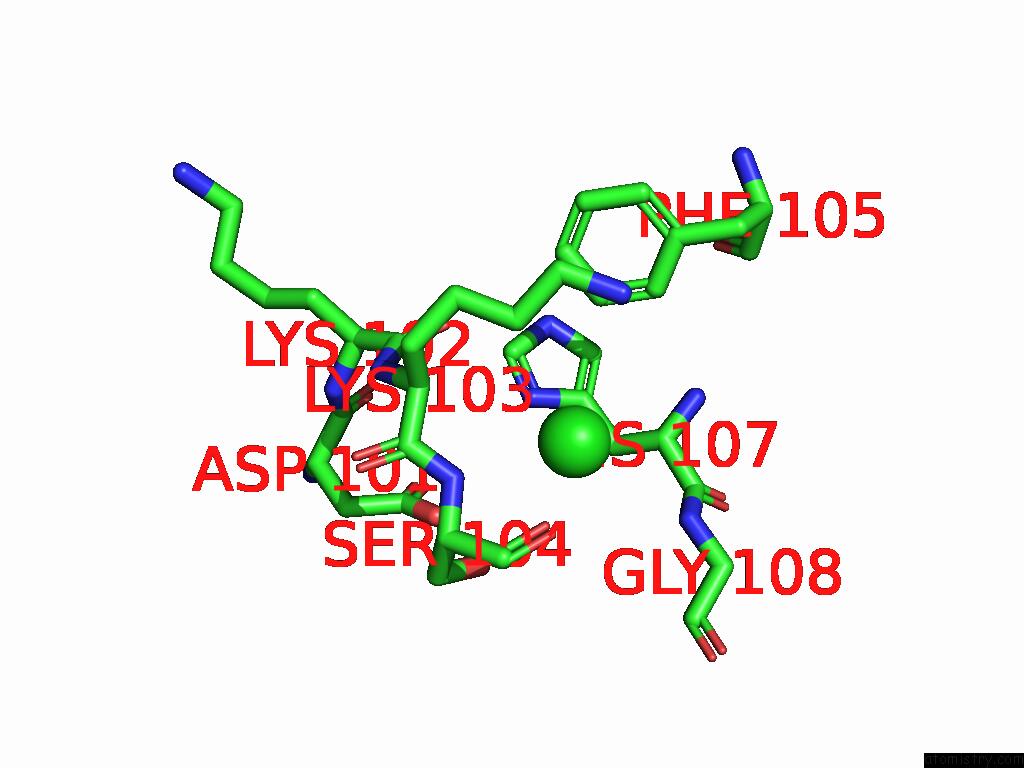
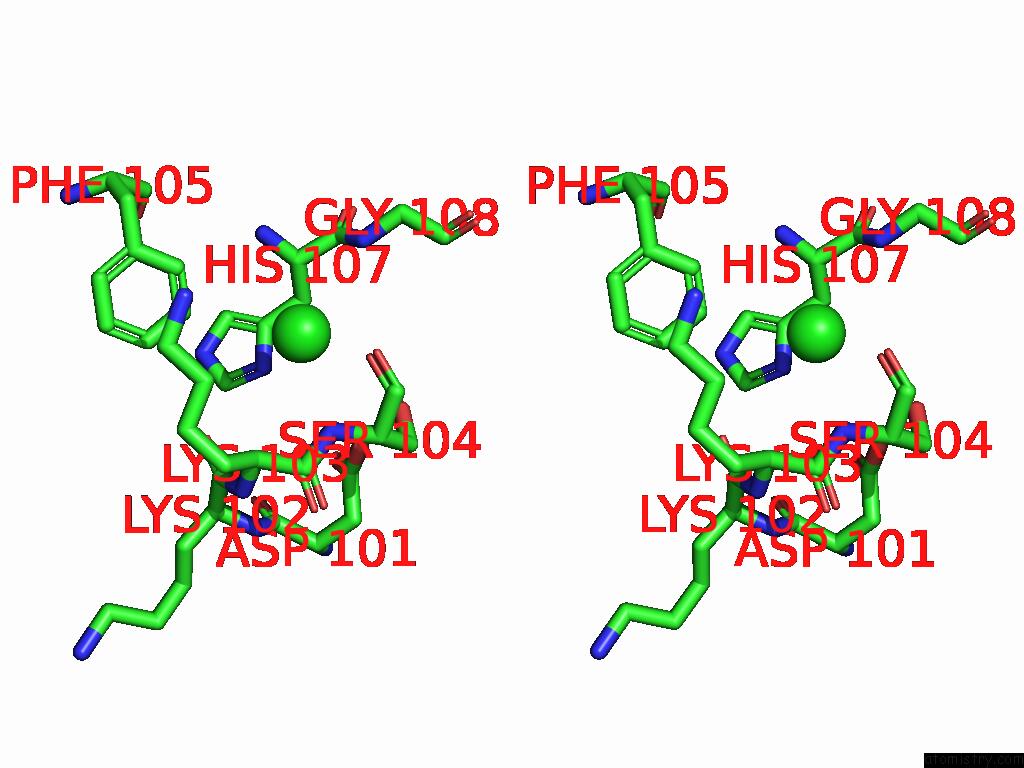
Chlorine binding site 4 out of 5 in 9ff2
Go back to
Chlorine binding site 4 out
of 5 in the Cryo-Em Structure of the BETA3 Homomeric Gaba(A) Receptor in Complex with Hsm in the Long-Lived Symmetric Desensitised State (C1)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Cryo-Em Structure of the BETA3 Homomeric Gaba(A) Receptor in Complex with Hsm in the Long-Lived Symmetric Desensitised State (C1) within 5.0Å range:
|
Chlorine binding site 5 out of 5 in 9ff2
Go back to
Chlorine binding site 5 out
of 5 in the Cryo-Em Structure of the BETA3 Homomeric Gaba(A) Receptor in Complex with Hsm in the Long-Lived Symmetric Desensitised State (C1)
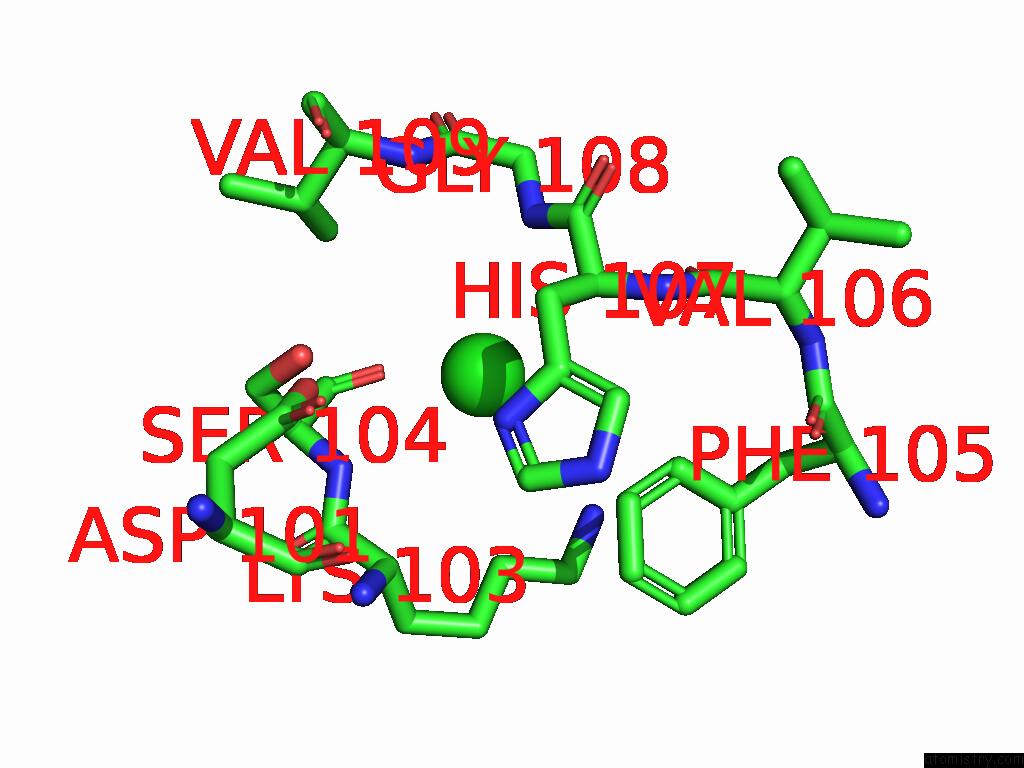
Mono view
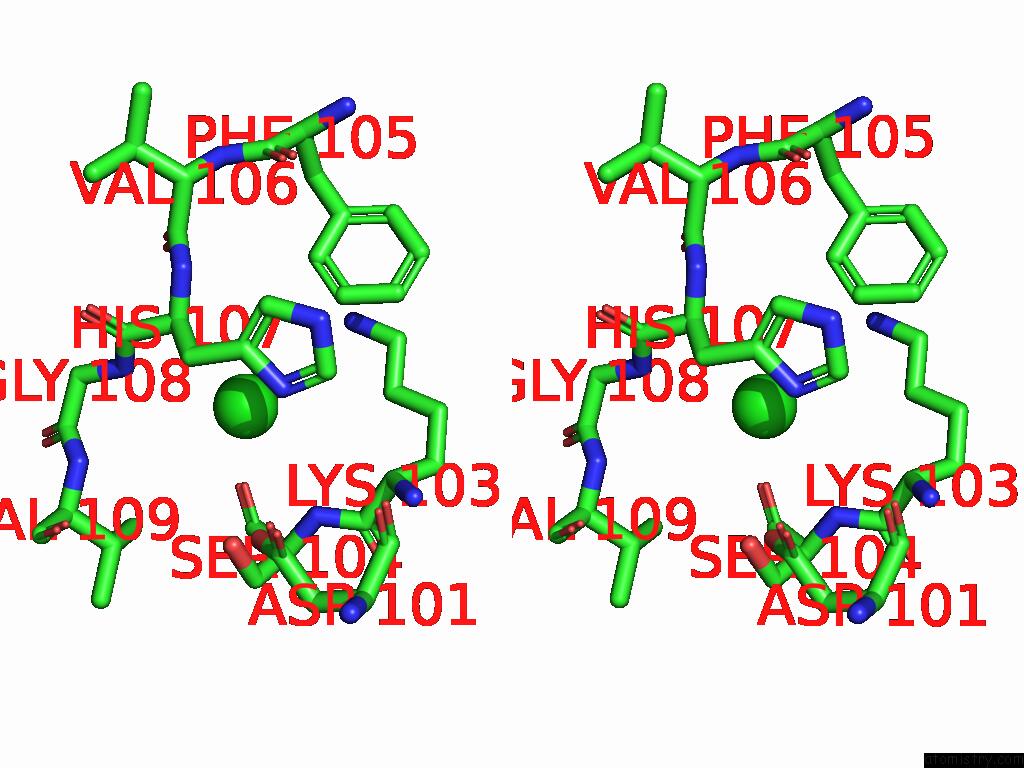
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Cryo-Em Structure of the BETA3 Homomeric Gaba(A) Receptor in Complex with Hsm in the Long-Lived Symmetric Desensitised State (C1) within 5.0Å range:
|
Reference:
D.B.Mihaylov,
T.Malinauskas,
A.R.Aricescu.
Cryo-Em Structure of the BETA3 Homomeric Gaba(A) Receptor in Complex with Hsm in the Long-Lived Symmetric Desensitised State (C1) To Be Published.
Page generated: Sun Jul 13 16:45:30 2025
Last articles
Cu in 1JUHCu in 1JVL
Cu in 1JRQ
Cu in 1JOI
Cu in 1JIF
Cu in 1JES
Cu in 1J9R
Cu in 1JER
Cu in 1JCV
Cu in 1ILU