Chlorine »
PDB 9fex-9g0g »
9fud »
Chlorine in PDB 9fud: Serial Microseconds Crystallography at ID29 Using Fixed-Target (Si Chip): Lysozyme - Without Ligand Glcnac (Apo)
Enzymatic activity of Serial Microseconds Crystallography at ID29 Using Fixed-Target (Si Chip): Lysozyme - Without Ligand Glcnac (Apo)
All present enzymatic activity of Serial Microseconds Crystallography at ID29 Using Fixed-Target (Si Chip): Lysozyme - Without Ligand Glcnac (Apo):
3.2.1.17;
3.2.1.17;
Protein crystallography data
The structure of Serial Microseconds Crystallography at ID29 Using Fixed-Target (Si Chip): Lysozyme - Without Ligand Glcnac (Apo), PDB code: 9fud
was solved by
J.Orlans,
S.L.Rose,
S.Basu,
D.De Sanctis,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 55.65 / 2.05 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 78.7, 78.7, 37.86, 90, 90, 90 |
| R / Rfree (%) | 21.6 / 25.2 |
Other elements in 9fud:
The structure of Serial Microseconds Crystallography at ID29 Using Fixed-Target (Si Chip): Lysozyme - Without Ligand Glcnac (Apo) also contains other interesting chemical elements:
| Sodium | (Na) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Serial Microseconds Crystallography at ID29 Using Fixed-Target (Si Chip): Lysozyme - Without Ligand Glcnac (Apo)
(pdb code 9fud). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 4 binding sites of Chlorine where determined in the Serial Microseconds Crystallography at ID29 Using Fixed-Target (Si Chip): Lysozyme - Without Ligand Glcnac (Apo), PDB code: 9fud:
Jump to Chlorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Chlorine where determined in the Serial Microseconds Crystallography at ID29 Using Fixed-Target (Si Chip): Lysozyme - Without Ligand Glcnac (Apo), PDB code: 9fud:
Jump to Chlorine binding site number: 1; 2; 3; 4;
Chlorine binding site 1 out of 4 in 9fud
Go back to
Chlorine binding site 1 out
of 4 in the Serial Microseconds Crystallography at ID29 Using Fixed-Target (Si Chip): Lysozyme - Without Ligand Glcnac (Apo)
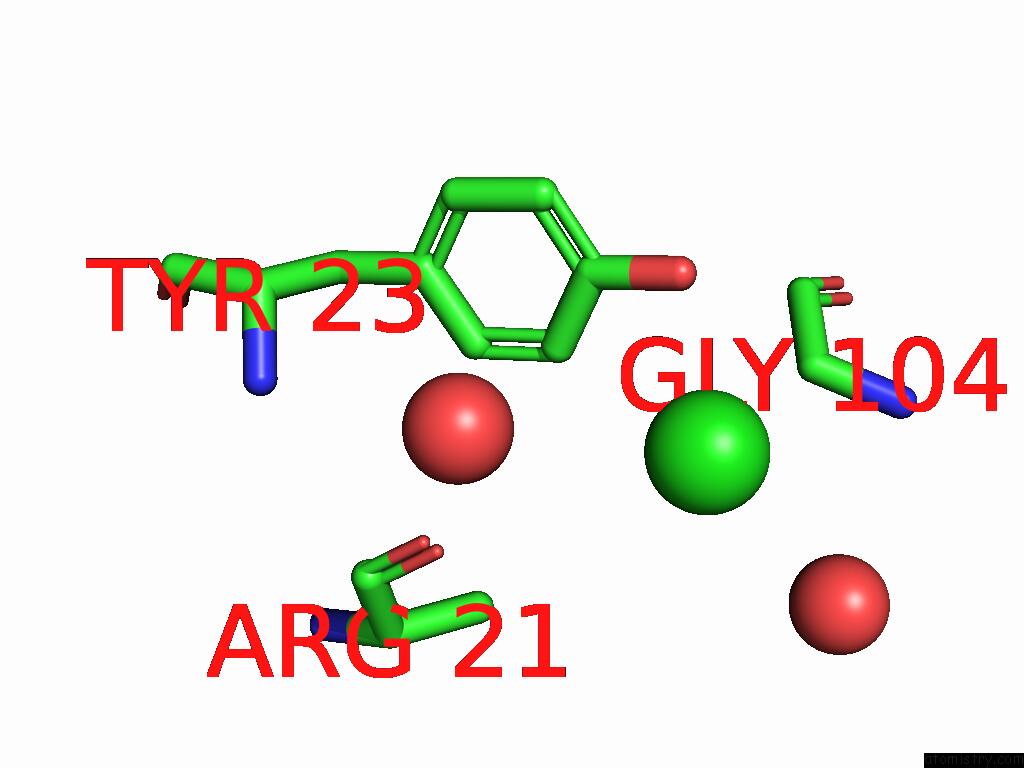
Mono view
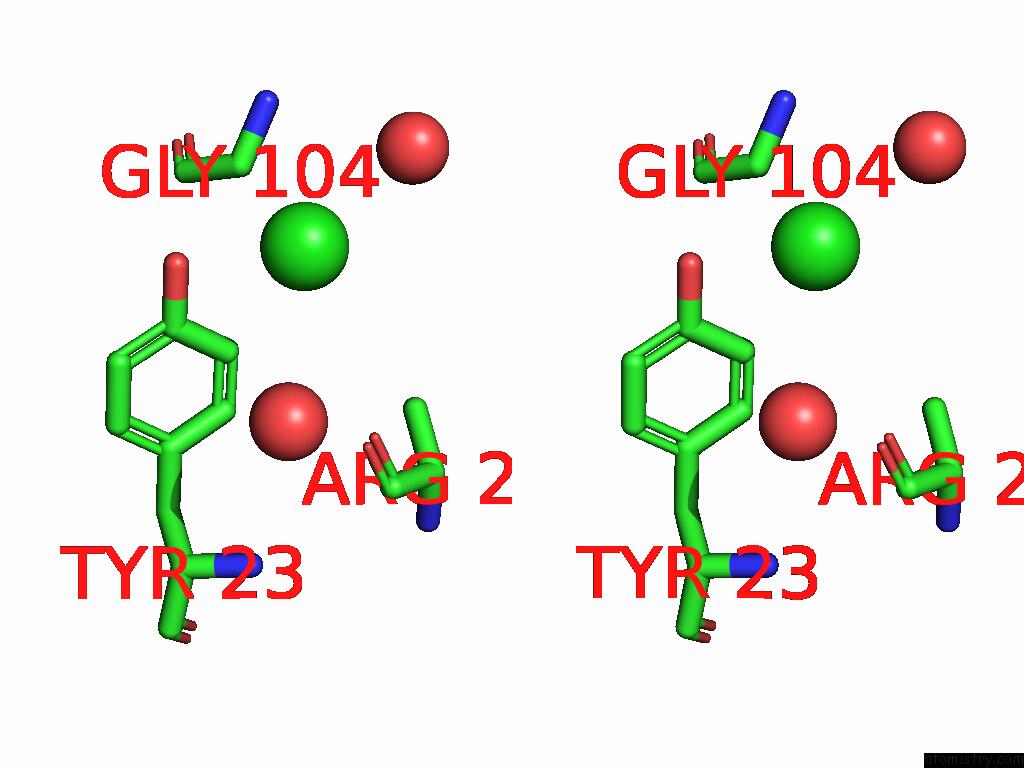
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Serial Microseconds Crystallography at ID29 Using Fixed-Target (Si Chip): Lysozyme - Without Ligand Glcnac (Apo) within 5.0Å range:
|
Chlorine binding site 2 out of 4 in 9fud
Go back to
Chlorine binding site 2 out
of 4 in the Serial Microseconds Crystallography at ID29 Using Fixed-Target (Si Chip): Lysozyme - Without Ligand Glcnac (Apo)
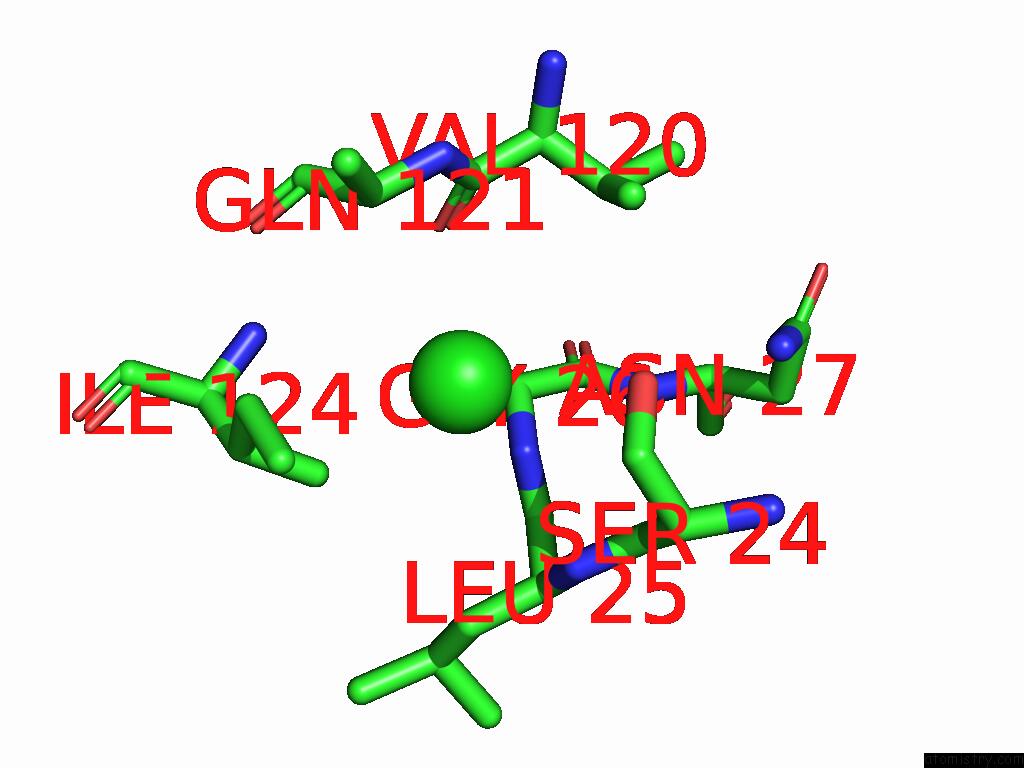
Mono view
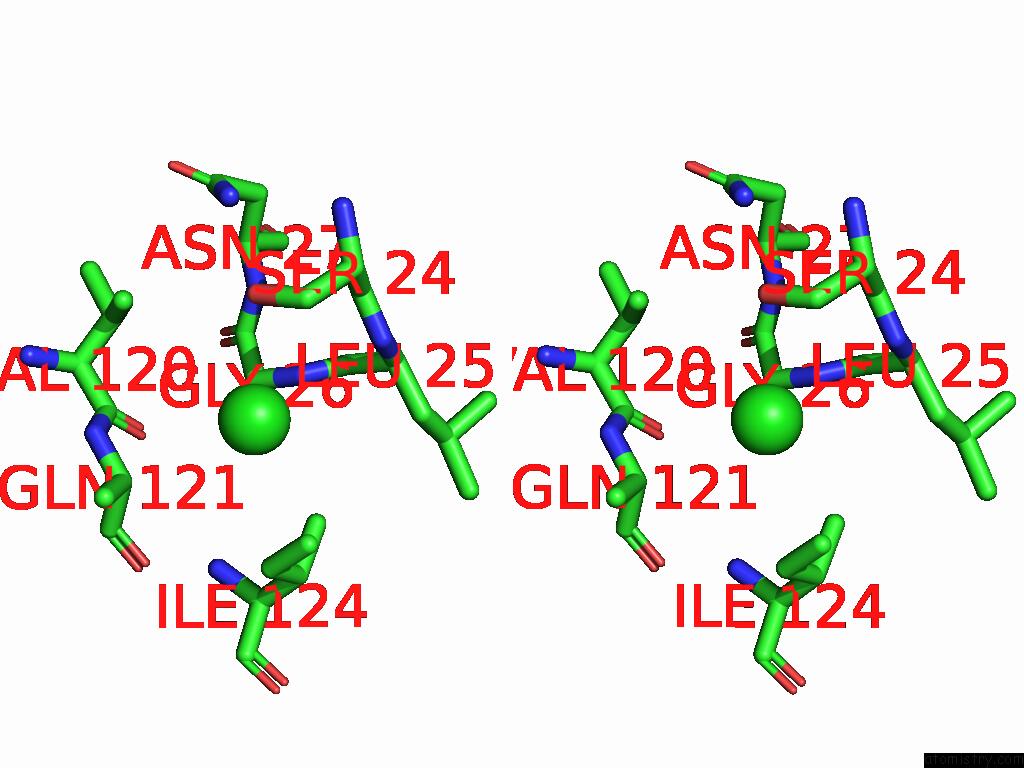
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Serial Microseconds Crystallography at ID29 Using Fixed-Target (Si Chip): Lysozyme - Without Ligand Glcnac (Apo) within 5.0Å range:
|
Chlorine binding site 3 out of 4 in 9fud
Go back to
Chlorine binding site 3 out
of 4 in the Serial Microseconds Crystallography at ID29 Using Fixed-Target (Si Chip): Lysozyme - Without Ligand Glcnac (Apo)
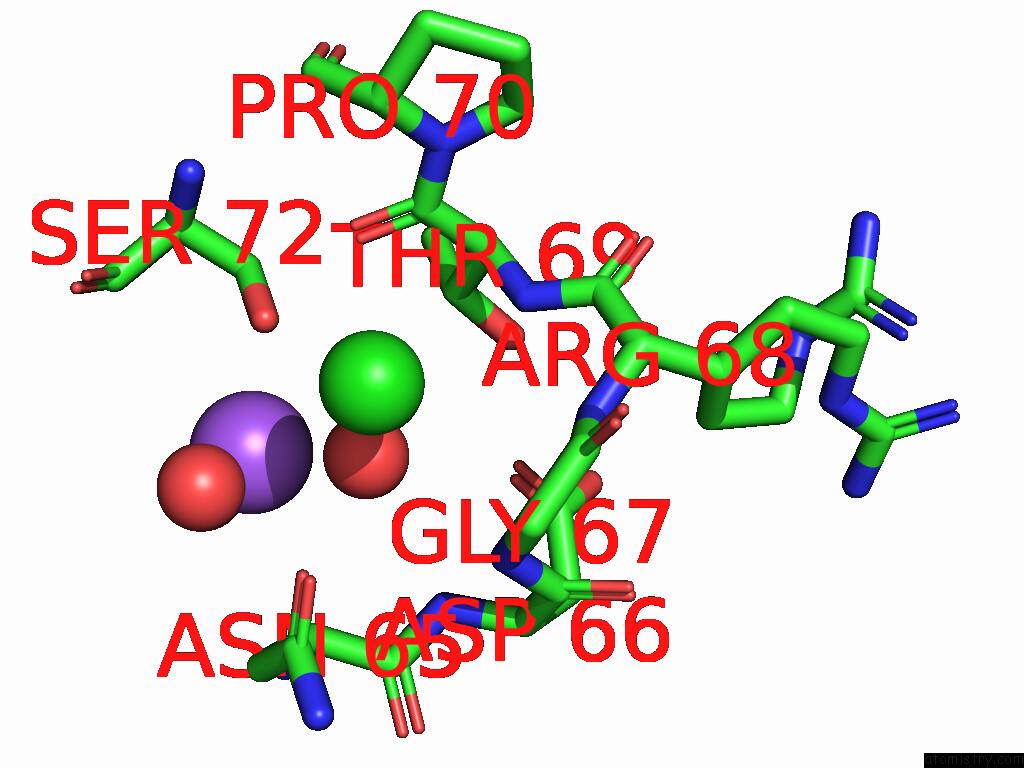
Mono view
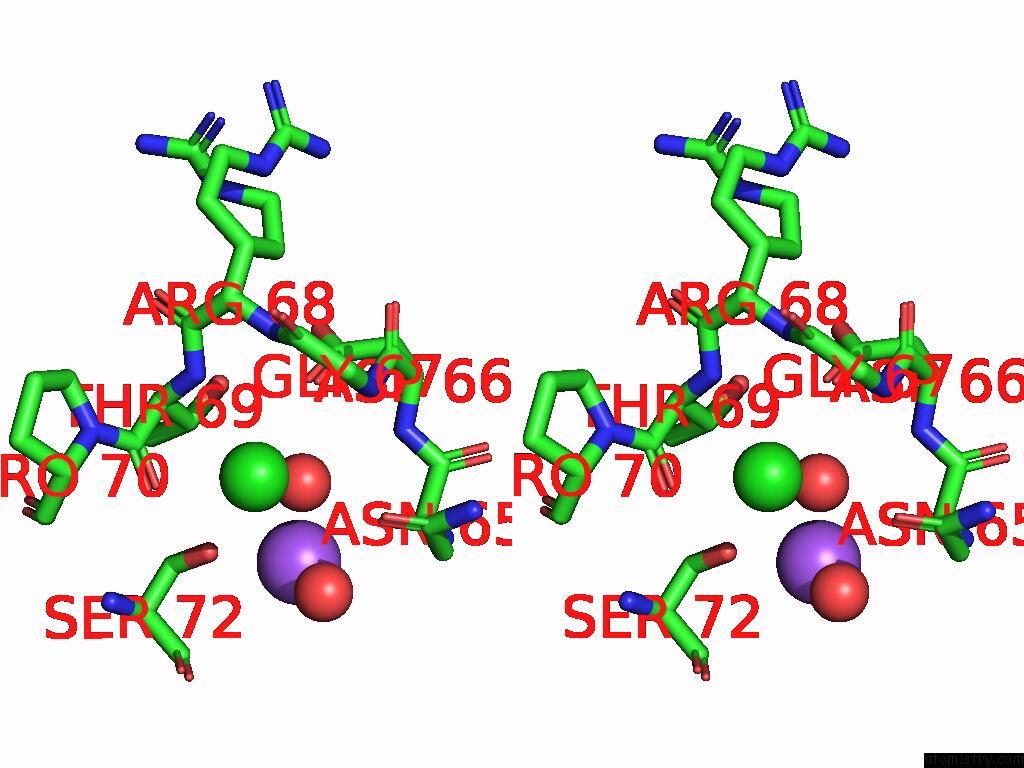
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Serial Microseconds Crystallography at ID29 Using Fixed-Target (Si Chip): Lysozyme - Without Ligand Glcnac (Apo) within 5.0Å range:
|
Chlorine binding site 4 out of 4 in 9fud
Go back to
Chlorine binding site 4 out
of 4 in the Serial Microseconds Crystallography at ID29 Using Fixed-Target (Si Chip): Lysozyme - Without Ligand Glcnac (Apo)
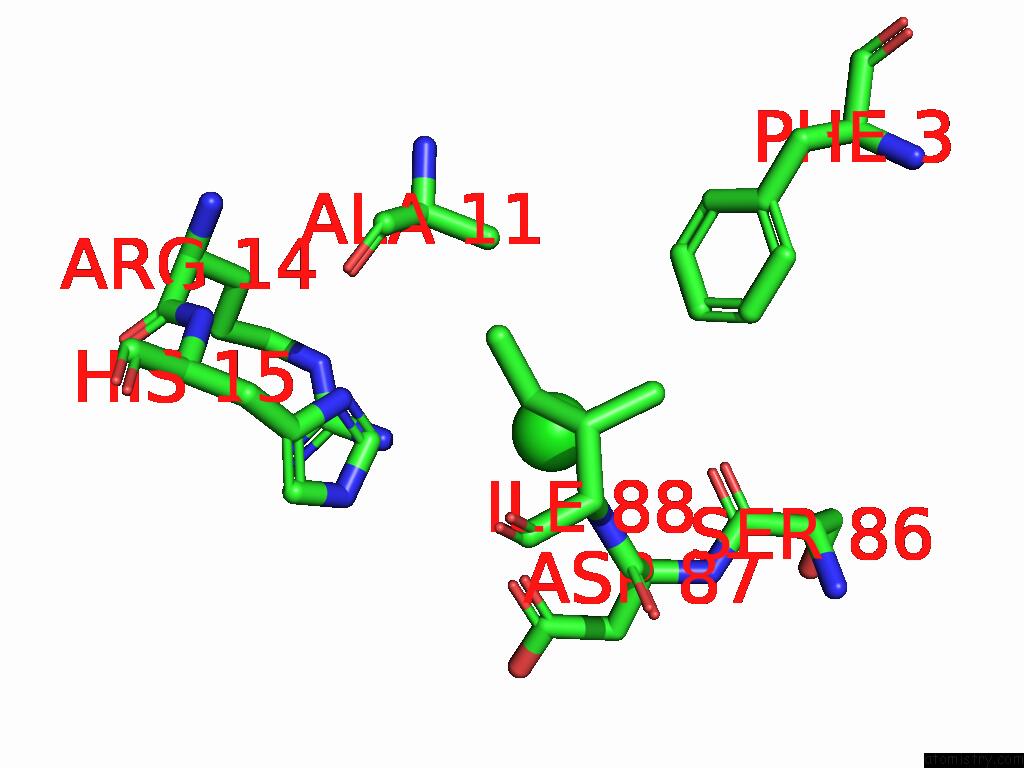
Mono view
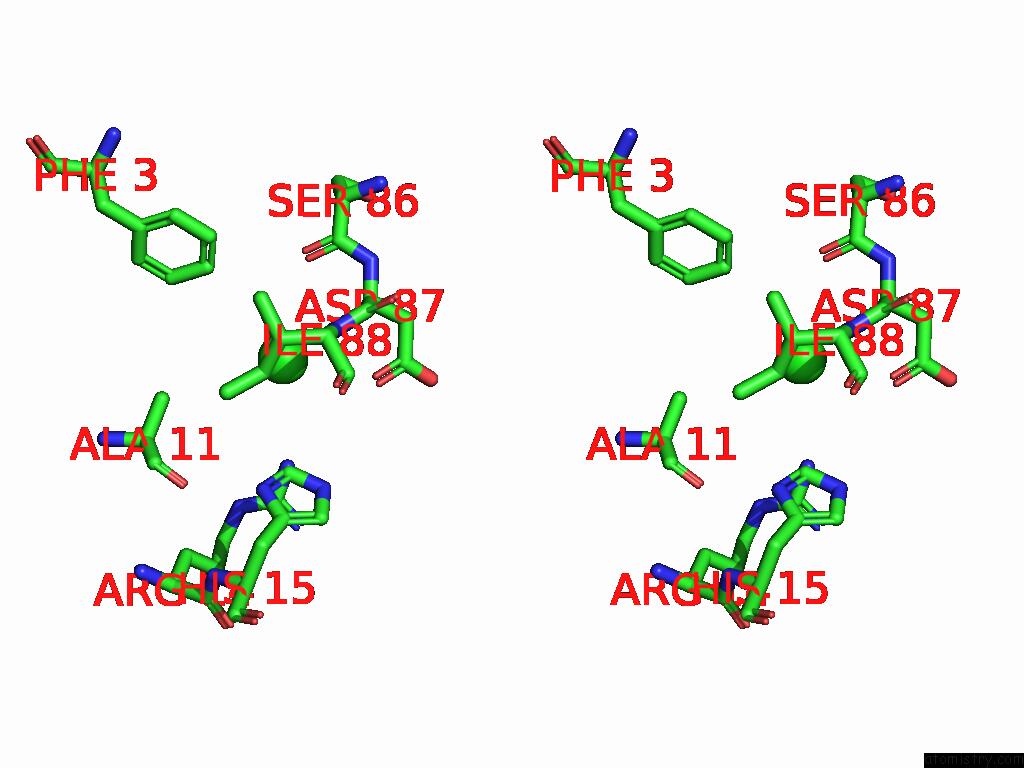
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Serial Microseconds Crystallography at ID29 Using Fixed-Target (Si Chip): Lysozyme - Without Ligand Glcnac (Apo) within 5.0Å range:
|
Reference:
J.Orlans,
S.L.Rose,
G.Ferguson,
M.Oscarsson,
A.Homs Puron,
A.Beteva,
S.Debionne,
P.Theveneau,
N.Coquelle,
J.Kieffer,
P.Busca,
J.Sinoir,
V.Armijo,
M.Lopez Marrero,
F.Felisaz,
G.Papp,
H.Gonzalez,
H.Caserotto,
F.Dobias,
J.Gigmes,
G.Lebon,
S.Basu,
D.De Sanctis.
Advancing Macromolecular Structure Determination with Microsecond X-Ray Pulses at A 4TH Generation Synchrotron. Commun Chem V. 8 6 2025.
ISSN: ESSN 2399-3669
PubMed: 39775172
DOI: 10.1038/S42004-024-01404-Y
Page generated: Sun Jul 13 16:50:46 2025
ISSN: ESSN 2399-3669
PubMed: 39775172
DOI: 10.1038/S42004-024-01404-Y
Last articles
F in 7MS5F in 7MRD
F in 7MS6
F in 7MR8
F in 7MRC
F in 7MPF
F in 7MR7
F in 7MPB
F in 7MR6
F in 7MNG