Chlorine »
PDB 9g0w-9h2d »
9g95 »
Chlorine in PDB 9g95: Lipid III Flippase Wzxe with NB10 Nanobody in Outward-Facing Conformation at 2.7552 A
Protein crystallography data
The structure of Lipid III Flippase Wzxe with NB10 Nanobody in Outward-Facing Conformation at 2.7552 A, PDB code: 9g95
was solved by
A.Le Bas,
K.El Omari,
M.Lee,
J.H.Naismith,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 44.78 / 2.80 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 271.54, 38.45, 56.96, 90, 93.09, 90 |
| R / Rfree (%) | 23.6 / 28.9 |
Other elements in 9g95:
The structure of Lipid III Flippase Wzxe with NB10 Nanobody in Outward-Facing Conformation at 2.7552 A also contains other interesting chemical elements:
| Zinc | (Zn) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Lipid III Flippase Wzxe with NB10 Nanobody in Outward-Facing Conformation at 2.7552 A
(pdb code 9g95). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Lipid III Flippase Wzxe with NB10 Nanobody in Outward-Facing Conformation at 2.7552 A, PDB code: 9g95:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Lipid III Flippase Wzxe with NB10 Nanobody in Outward-Facing Conformation at 2.7552 A, PDB code: 9g95:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 9g95
Go back to
Chlorine binding site 1 out
of 2 in the Lipid III Flippase Wzxe with NB10 Nanobody in Outward-Facing Conformation at 2.7552 A
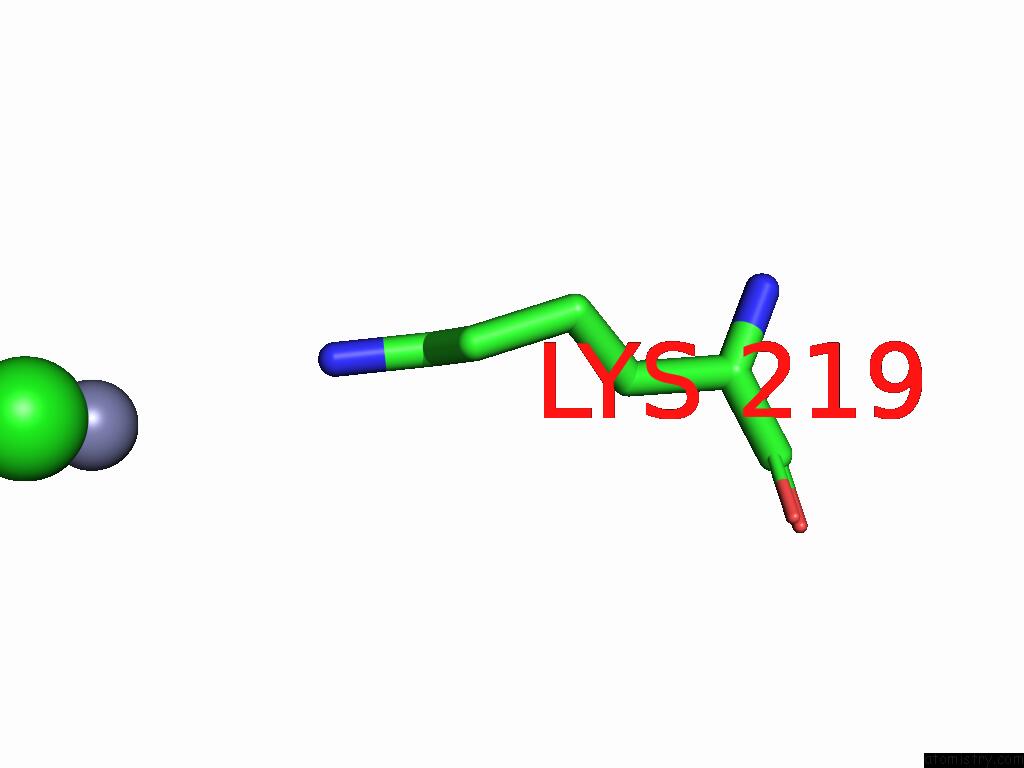
Mono view
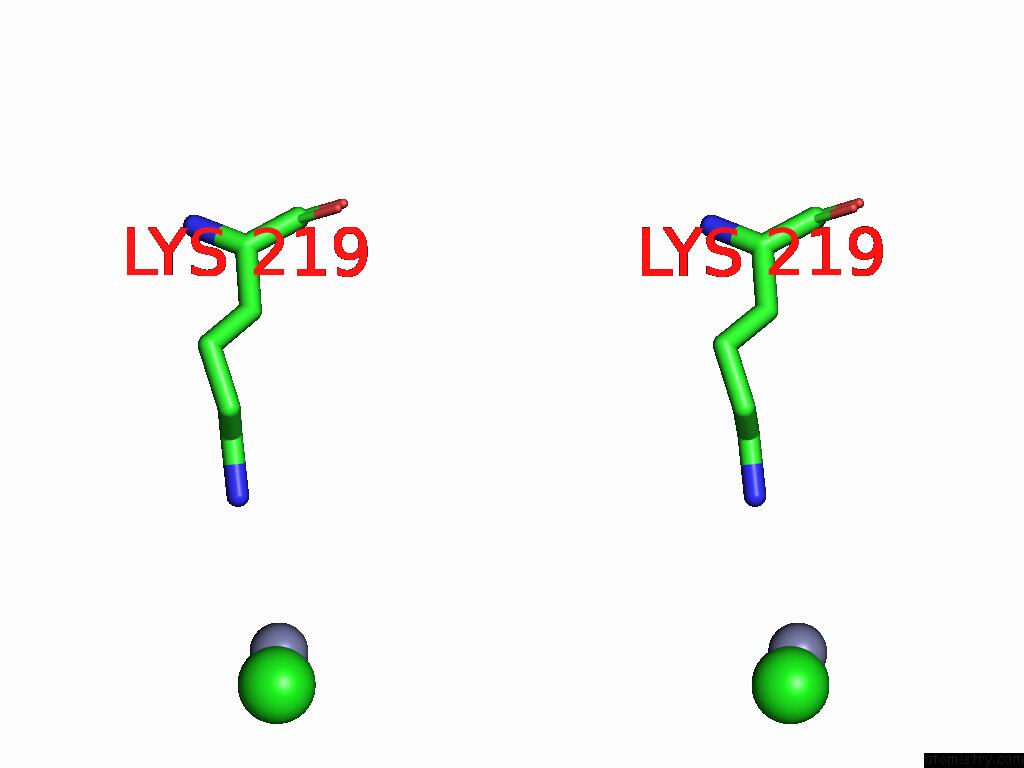
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Lipid III Flippase Wzxe with NB10 Nanobody in Outward-Facing Conformation at 2.7552 A within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 9g95
Go back to
Chlorine binding site 2 out
of 2 in the Lipid III Flippase Wzxe with NB10 Nanobody in Outward-Facing Conformation at 2.7552 A
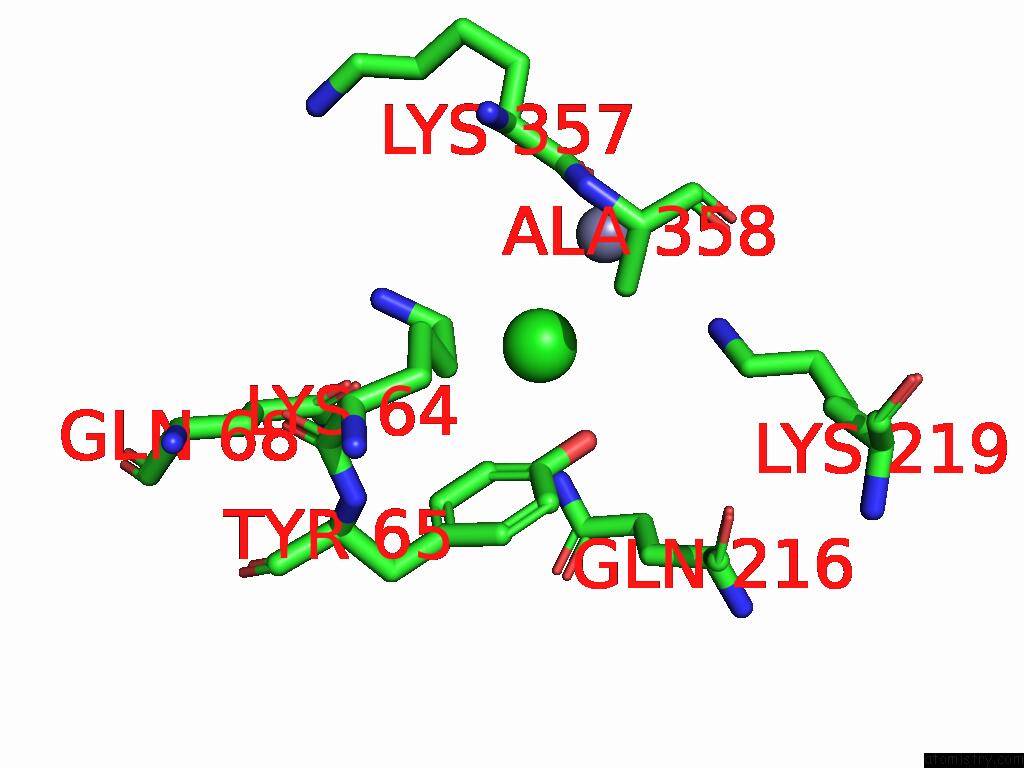
Mono view
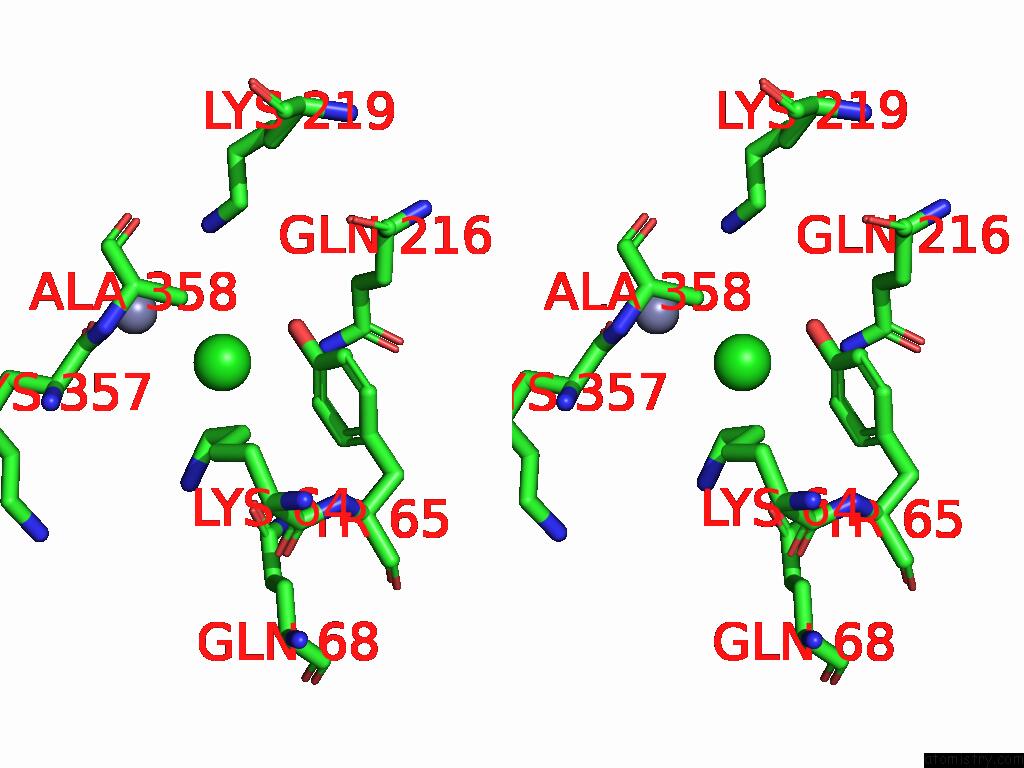
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Lipid III Flippase Wzxe with NB10 Nanobody in Outward-Facing Conformation at 2.7552 A within 5.0Å range:
|
Reference:
A.Le Bas,
B.R.Clarke,
T.Teelucksingh,
M.Lee,
K.El Omari,
A.M.Giltrap,
S.A.Mcmahon,
H.Liu,
J.H.Beale,
V.Mykhaylyk,
R.Duman,
N.G.Paterson,
P.N.Ward,
P.J.Harrison,
M.Weckener,
E.Pardon,
J.Steyaert,
H.Liu,
A.Quigley,
B.G.Davis,
A.Wagner,
C.Whitfield,
J.H.Naismith.
Structure of Wzxe the Lipid III Flippase For Enterobacterial Common Antigen Polysaccharide. Open Biology V. 15 40310 2025.
ISSN: ESSN 2046-2441
PubMed: 39772807
DOI: 10.1098/RSOB.240310
Page generated: Sun Jul 13 16:55:14 2025
ISSN: ESSN 2046-2441
PubMed: 39772807
DOI: 10.1098/RSOB.240310
Last articles
F in 7PRMF in 7PQV
F in 7PQS
F in 7PMM
F in 7POL
F in 7POR
F in 7PNS
F in 7P81
F in 7PLI
F in 7PJR