Chlorine »
PDB 1lwk-1mj5 »
1m5k »
Chlorine in PDB 1m5k: Crystal Structure of A Hairpin Ribozyme in the Catalytically-Active Conformation
Protein crystallography data
The structure of Crystal Structure of A Hairpin Ribozyme in the Catalytically-Active Conformation, PDB code: 1m5k
was solved by
P.B.Rupert,
A.R.Ferre-D'amare,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 61.07 / 2.40 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 256.300, 44.100, 102.400, 90.00, 109.00, 90.00 |
| R / Rfree (%) | 22.9 / 28.4 |
Other elements in 1m5k:
The structure of Crystal Structure of A Hairpin Ribozyme in the Catalytically-Active Conformation also contains other interesting chemical elements:
| Iodine | (I) | 2 atoms |
| Calcium | (Ca) | 33 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of A Hairpin Ribozyme in the Catalytically-Active Conformation
(pdb code 1m5k). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Crystal Structure of A Hairpin Ribozyme in the Catalytically-Active Conformation, PDB code: 1m5k:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Crystal Structure of A Hairpin Ribozyme in the Catalytically-Active Conformation, PDB code: 1m5k:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 1m5k
Go back to
Chlorine binding site 1 out
of 2 in the Crystal Structure of A Hairpin Ribozyme in the Catalytically-Active Conformation
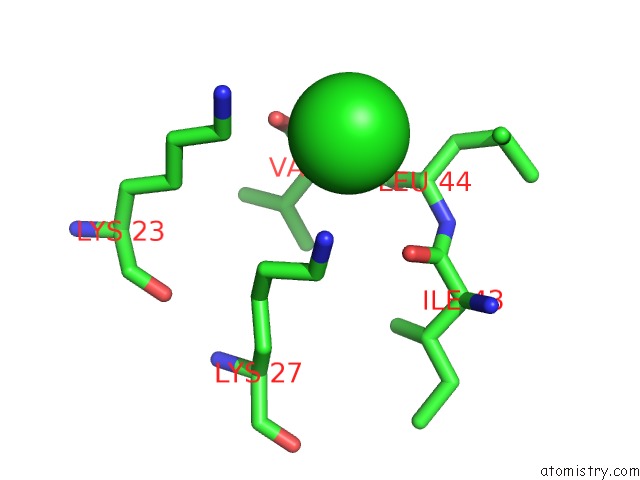
Mono view
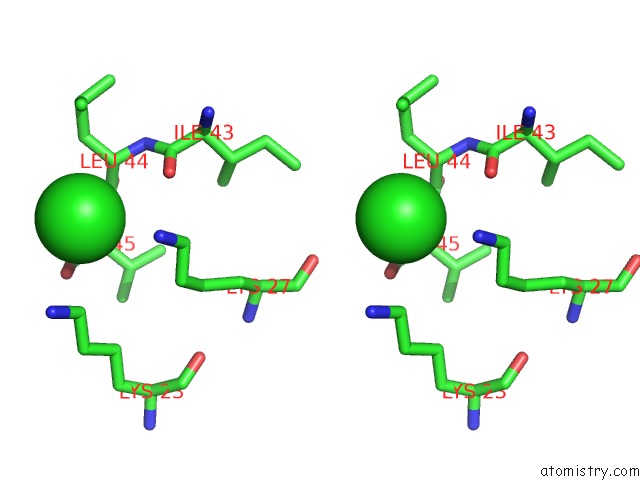
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of A Hairpin Ribozyme in the Catalytically-Active Conformation within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 1m5k
Go back to
Chlorine binding site 2 out
of 2 in the Crystal Structure of A Hairpin Ribozyme in the Catalytically-Active Conformation
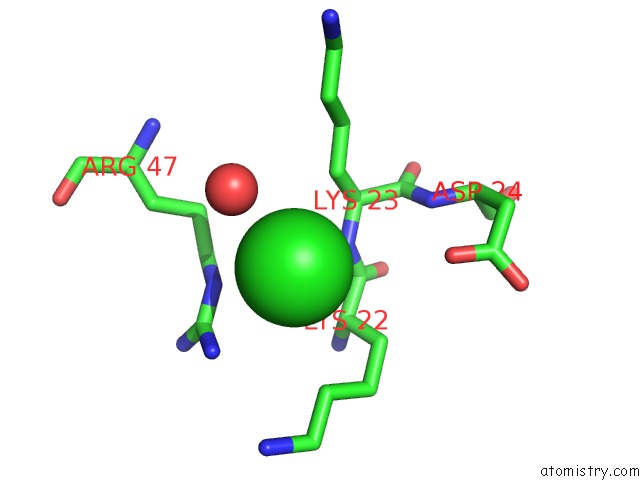
Mono view
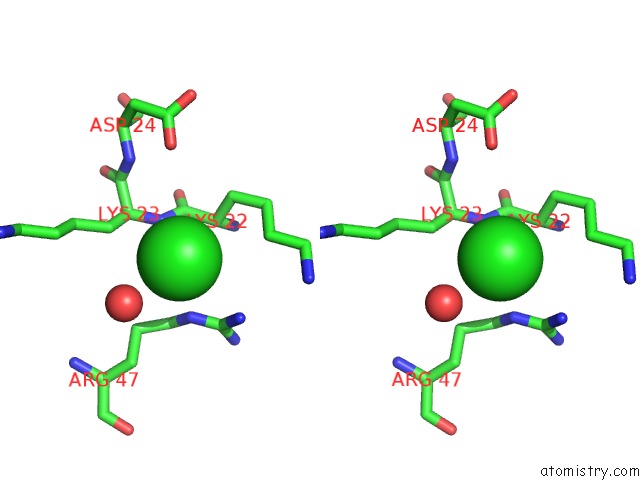
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of A Hairpin Ribozyme in the Catalytically-Active Conformation within 5.0Å range:
|
Reference:
P.B.Rupert,
A.P.Massey,
S.T.Sigurdsson,
A.R.Ferre-D'amare.
Transition State Stabilization By A Catalytic Rna Science V. 298 1421 2002.
ISSN: ISSN 0036-8075
PubMed: 11298439
DOI: 10.1126/SCIENCE.1076093
Page generated: Thu Jul 10 18:09:08 2025
ISSN: ISSN 0036-8075
PubMed: 11298439
DOI: 10.1126/SCIENCE.1076093
Last articles
F in 5CMKF in 5CRZ
F in 5CP5
F in 5CI3
F in 5CI2
F in 5CI1
F in 5CI0
F in 5CGQ
F in 5CDT
F in 5CEP