Chlorine »
PDB 1mkp-1nc4 »
1mwq »
Chlorine in PDB 1mwq: Structure of HI0828, A Hypothetical Protein From Haemophilus Influenzae with A Putative Active-Site Phosphohistidine
Protein crystallography data
The structure of Structure of HI0828, A Hypothetical Protein From Haemophilus Influenzae with A Putative Active-Site Phosphohistidine, PDB code: 1mwq
was solved by
M.A.Willis,
W.Krajewski,
V.R.Chalamasetty,
P.Reddy,
A.Howard,
O.Herzberg,
Structure 2 Function Project (S2F),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 15.00 / 0.99 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 42.476, 63.317, 75.465, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 10.8 / 13.2 |
Other elements in 1mwq:
The structure of Structure of HI0828, A Hypothetical Protein From Haemophilus Influenzae with A Putative Active-Site Phosphohistidine also contains other interesting chemical elements:
| Arsenic | (As) | 1 atom |
| Zinc | (Zn) | 4 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Structure of HI0828, A Hypothetical Protein From Haemophilus Influenzae with A Putative Active-Site Phosphohistidine
(pdb code 1mwq). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 6 binding sites of Chlorine where determined in the Structure of HI0828, A Hypothetical Protein From Haemophilus Influenzae with A Putative Active-Site Phosphohistidine, PDB code: 1mwq:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Chlorine where determined in the Structure of HI0828, A Hypothetical Protein From Haemophilus Influenzae with A Putative Active-Site Phosphohistidine, PDB code: 1mwq:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
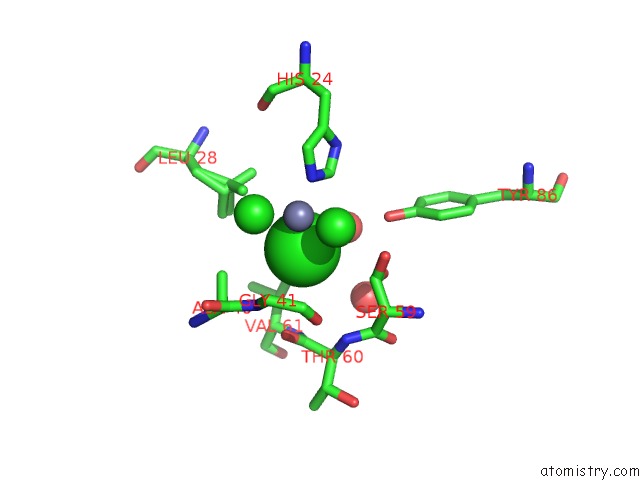
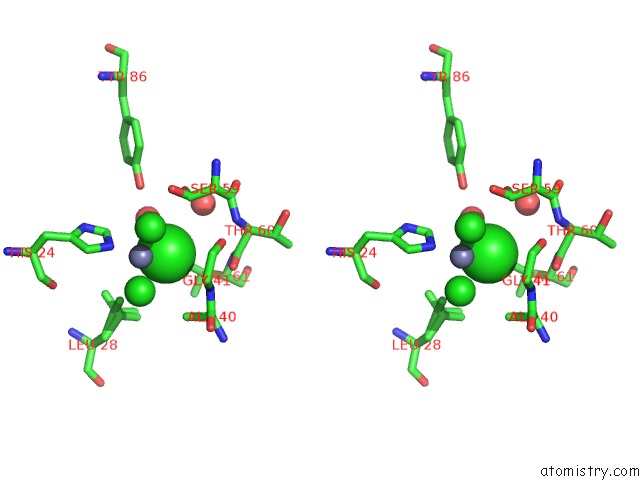
Chlorine binding site 1 out of 6 in 1mwq
Go back to
Chlorine binding site 1 out
of 6 in the Structure of HI0828, A Hypothetical Protein From Haemophilus Influenzae with A Putative Active-Site Phosphohistidine
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Structure of HI0828, A Hypothetical Protein From Haemophilus Influenzae with A Putative Active-Site Phosphohistidine within 5.0Å range:
|
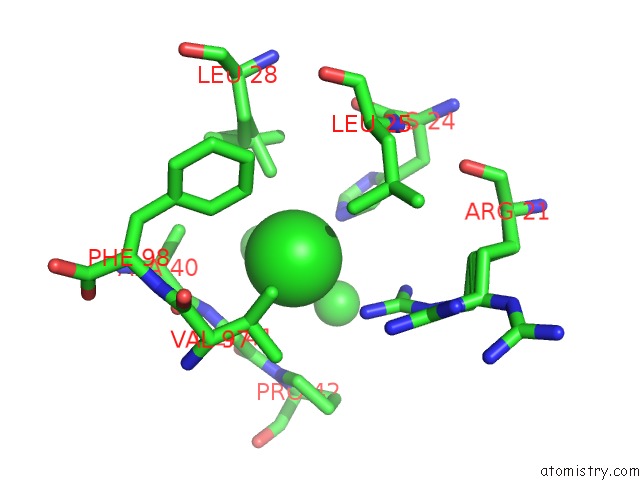
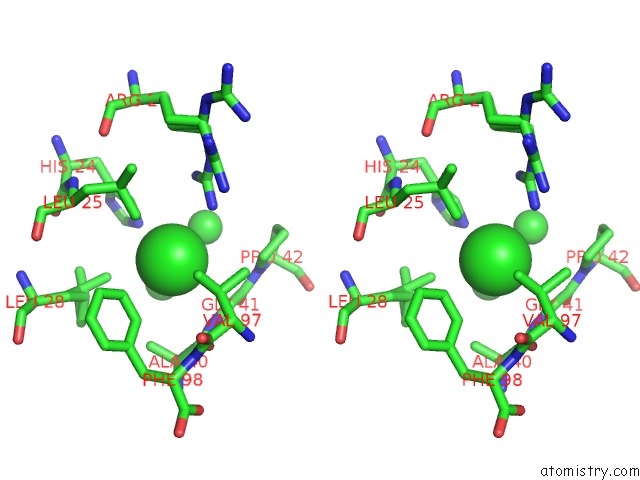
Chlorine binding site 2 out of 6 in 1mwq
Go back to
Chlorine binding site 2 out
of 6 in the Structure of HI0828, A Hypothetical Protein From Haemophilus Influenzae with A Putative Active-Site Phosphohistidine
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Structure of HI0828, A Hypothetical Protein From Haemophilus Influenzae with A Putative Active-Site Phosphohistidine within 5.0Å range:
|
Chlorine binding site 3 out of 6 in 1mwq
Go back to
Chlorine binding site 3 out
of 6 in the Structure of HI0828, A Hypothetical Protein From Haemophilus Influenzae with A Putative Active-Site Phosphohistidine
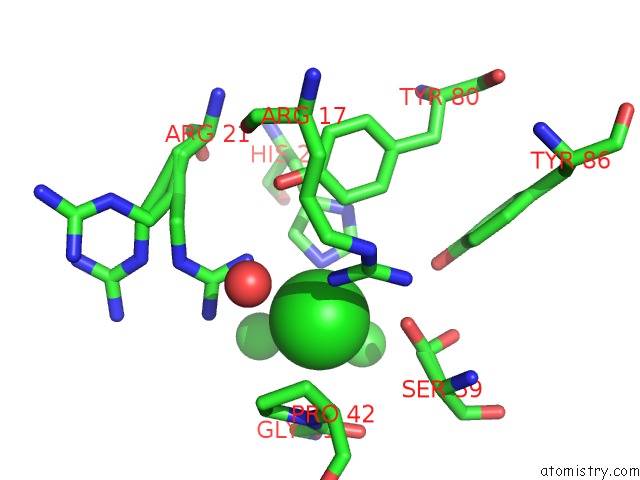
Mono view
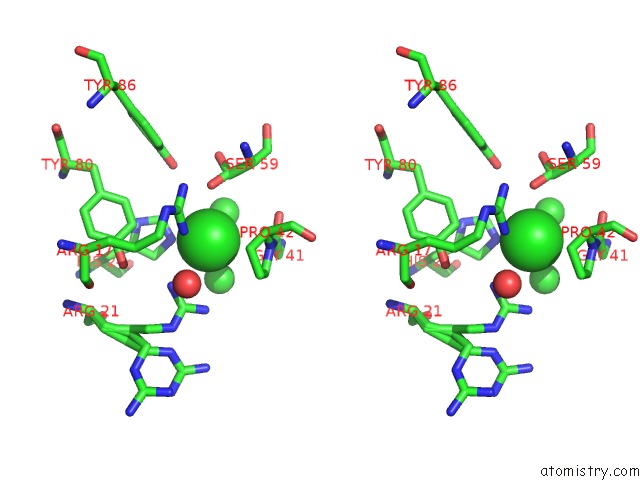
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Structure of HI0828, A Hypothetical Protein From Haemophilus Influenzae with A Putative Active-Site Phosphohistidine within 5.0Å range:
|
Chlorine binding site 4 out of 6 in 1mwq
Go back to
Chlorine binding site 4 out
of 6 in the Structure of HI0828, A Hypothetical Protein From Haemophilus Influenzae with A Putative Active-Site Phosphohistidine
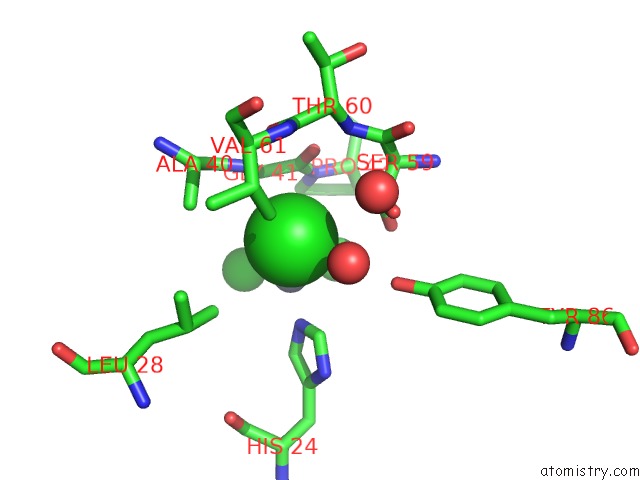
Mono view
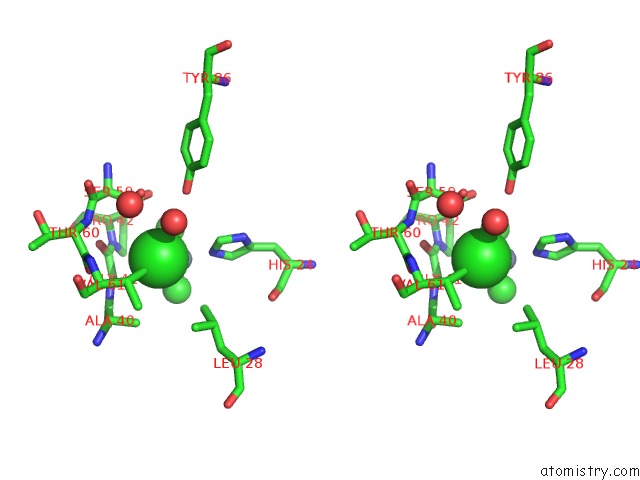
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Structure of HI0828, A Hypothetical Protein From Haemophilus Influenzae with A Putative Active-Site Phosphohistidine within 5.0Å range:
|
Chlorine binding site 5 out of 6 in 1mwq
Go back to
Chlorine binding site 5 out
of 6 in the Structure of HI0828, A Hypothetical Protein From Haemophilus Influenzae with A Putative Active-Site Phosphohistidine
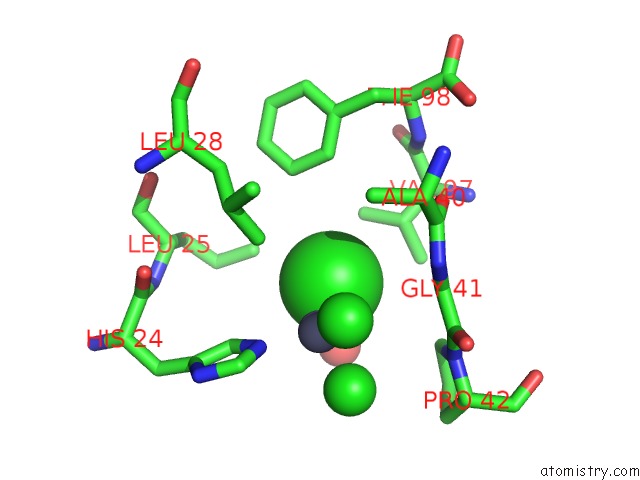
Mono view
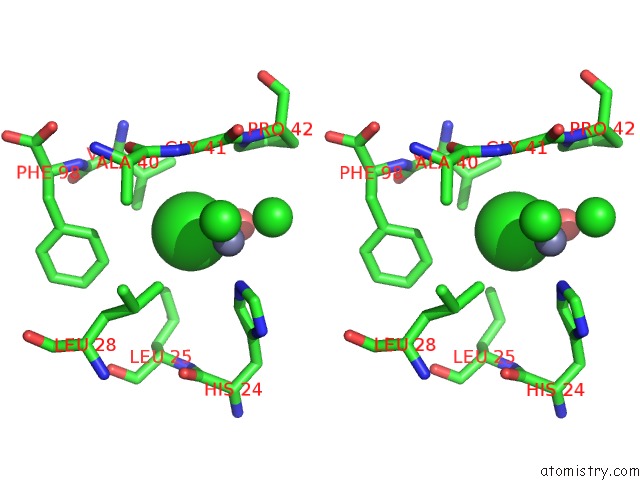
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Structure of HI0828, A Hypothetical Protein From Haemophilus Influenzae with A Putative Active-Site Phosphohistidine within 5.0Å range:
|
Chlorine binding site 6 out of 6 in 1mwq
Go back to
Chlorine binding site 6 out
of 6 in the Structure of HI0828, A Hypothetical Protein From Haemophilus Influenzae with A Putative Active-Site Phosphohistidine
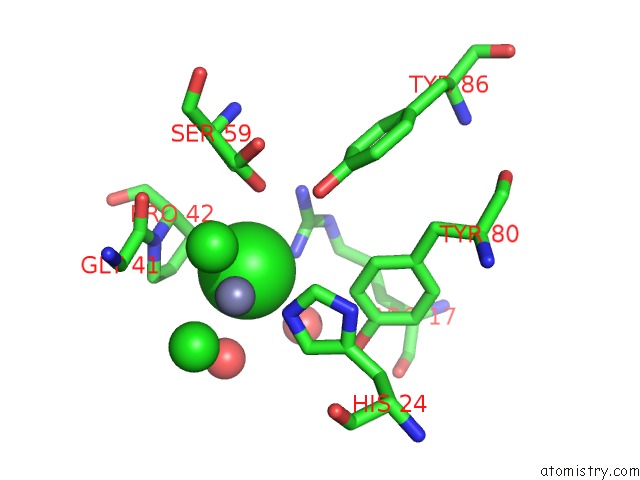
Mono view
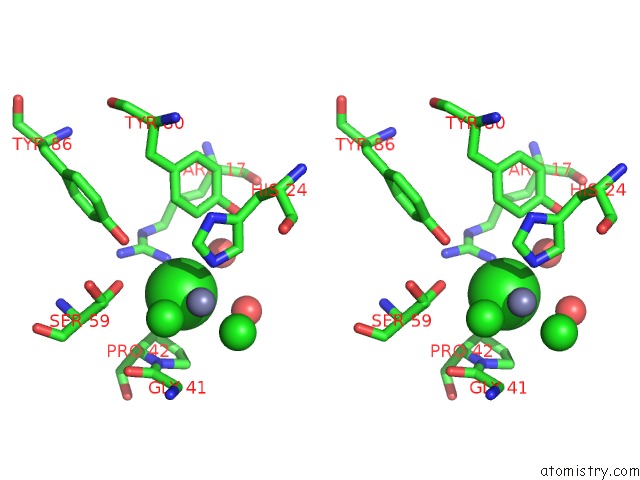
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Structure of HI0828, A Hypothetical Protein From Haemophilus Influenzae with A Putative Active-Site Phosphohistidine within 5.0Å range:
|
Reference:
M.A.Willis,
F.Song,
Z.Zhuang,
W.Krajewski,
V.R.Chalamasetty,
P.Reddy,
A.Howard,
D.Dunaway-Mariano,
O.Herzberg.
Structure of Ycii From Haemophilus Influenzae (HI0828) Reveals A Ferredoxin-Like Alpha/Beta-Fold with A Histidine/Aspartate Centered Catalytic Site Proteins V. 59 648 2005.
ISSN: ISSN 0887-3585
PubMed: 15779043
DOI: 10.1002/PROT.20411
Page generated: Thu Jul 10 18:16:09 2025
ISSN: ISSN 0887-3585
PubMed: 15779043
DOI: 10.1002/PROT.20411
Last articles
K in 5E33K in 5E2Q
K in 5E1A
K in 5DWX
K in 5DUN
K in 5DSX
K in 5DRY
K in 5DRT
K in 5DMJ
K in 5DQK