Chlorine »
PDB 1ti7-1u33 »
1tzm »
Chlorine in PDB 1tzm: Crystal Structure of Acc Deaminase Complexed with Substrate Analog B- Chloro-D-Alanine
Enzymatic activity of Crystal Structure of Acc Deaminase Complexed with Substrate Analog B- Chloro-D-Alanine
All present enzymatic activity of Crystal Structure of Acc Deaminase Complexed with Substrate Analog B- Chloro-D-Alanine:
3.5.99.7;
3.5.99.7;
Protein crystallography data
The structure of Crystal Structure of Acc Deaminase Complexed with Substrate Analog B- Chloro-D-Alanine, PDB code: 1tzm
was solved by
S.Karthikeyan,
Q.Zhou,
Z.Zhao,
C.L.Kao,
Z.Tao,
H.Robinson,
H.W.Liu,
H.Zhang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.86 / 2.08 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 67.640, 68.263, 349.748, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 22.6 / 26.8 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Acc Deaminase Complexed with Substrate Analog B- Chloro-D-Alanine
(pdb code 1tzm). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Crystal Structure of Acc Deaminase Complexed with Substrate Analog B- Chloro-D-Alanine, PDB code: 1tzm:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Crystal Structure of Acc Deaminase Complexed with Substrate Analog B- Chloro-D-Alanine, PDB code: 1tzm:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 1tzm
Go back to
Chlorine binding site 1 out
of 2 in the Crystal Structure of Acc Deaminase Complexed with Substrate Analog B- Chloro-D-Alanine
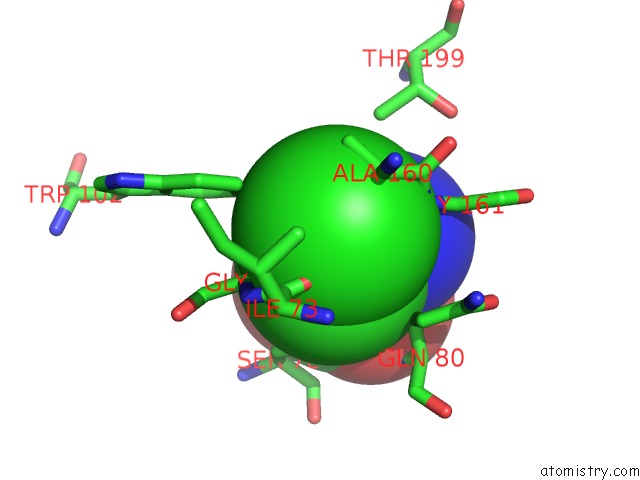
Mono view
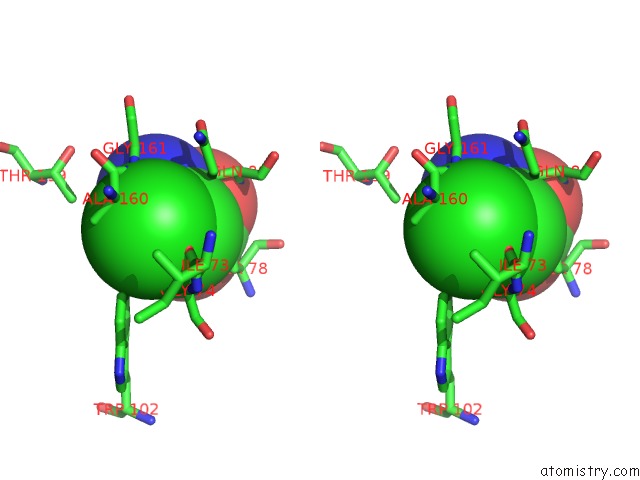
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Acc Deaminase Complexed with Substrate Analog B- Chloro-D-Alanine within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 1tzm
Go back to
Chlorine binding site 2 out
of 2 in the Crystal Structure of Acc Deaminase Complexed with Substrate Analog B- Chloro-D-Alanine
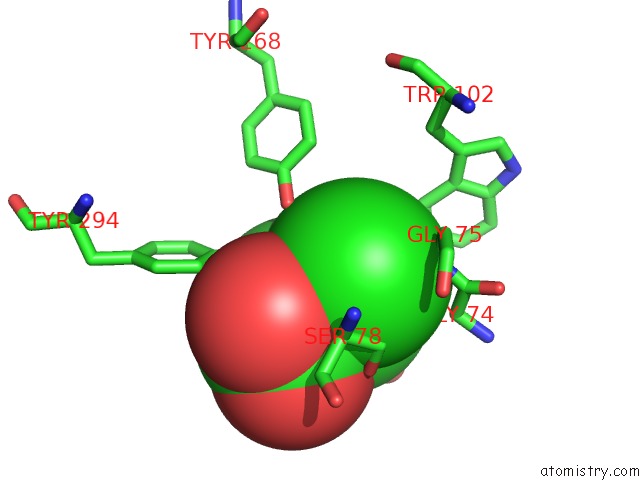
Mono view
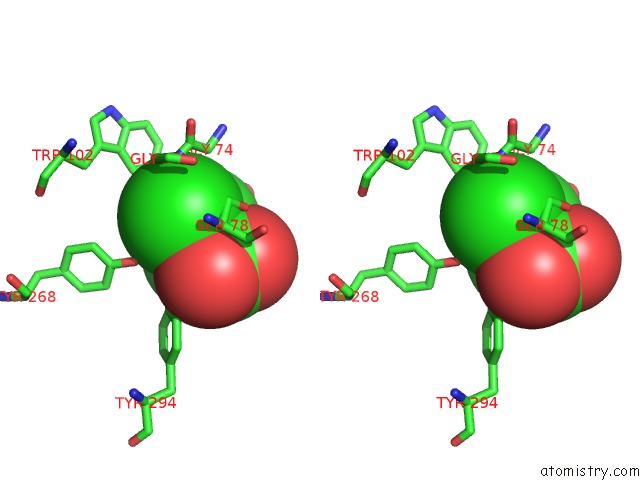
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Acc Deaminase Complexed with Substrate Analog B- Chloro-D-Alanine within 5.0Å range:
|
Reference:
S.Karthikeyan,
Q.Zhou,
Z.Zhao,
C.L.Kao,
Z.Tao,
H.Robinson,
H.W.Liu,
H.Zhang.
Structural Analysis of Pseudomonas 1-Aminocyclopropane-1-Carboxylate Deaminase Complexes: Insight Into the Mechanism of A Unique Pyridoxal-5'-Phosphate Dependent Cyclopropane Ring-Opening Reaction Biochemistry V. 43 13328 2004.
ISSN: ISSN 0006-2960
PubMed: 15491139
DOI: 10.1021/BI048878G
Page generated: Thu Jul 10 19:39:50 2025
ISSN: ISSN 0006-2960
PubMed: 15491139
DOI: 10.1021/BI048878G
Last articles
I in 8A25I in 7WYJ
I in 7YWW
I in 8A24
I in 7Z76
I in 7YDX
I in 7XME
I in 7YUZ
I in 7XMK
I in 7XC1