Chlorine »
PDB 1wvu-1xkk »
1xkk »
Chlorine in PDB 1xkk: Egfr Kinase Domain Complexed with A Quinazoline Inhibitor- GW572016
Enzymatic activity of Egfr Kinase Domain Complexed with A Quinazoline Inhibitor- GW572016
All present enzymatic activity of Egfr Kinase Domain Complexed with A Quinazoline Inhibitor- GW572016:
2.7.1.112;
2.7.1.112;
Protein crystallography data
The structure of Egfr Kinase Domain Complexed with A Quinazoline Inhibitor- GW572016, PDB code: 1xkk
was solved by
E.R.Wood,
A.T.Truesdale,
O.B.Mcdonald,
D.Yuan,
A.Hassell,
S.H.Dickerson,
B.Ellis,
C.Pennisi,
E.Horne,
K.Lackey,
K.J.Alligood,
D.W.Rusnak,
T.M.Gilmer,
L.M.Shewchuk,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 35.44 / 2.40 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 45.653, 67.144, 102.880, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.9 / 25.5 |
Other elements in 1xkk:
The structure of Egfr Kinase Domain Complexed with A Quinazoline Inhibitor- GW572016 also contains other interesting chemical elements:
| Fluorine | (F) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Egfr Kinase Domain Complexed with A Quinazoline Inhibitor- GW572016
(pdb code 1xkk). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Egfr Kinase Domain Complexed with A Quinazoline Inhibitor- GW572016, PDB code: 1xkk:
In total only one binding site of Chlorine was determined in the Egfr Kinase Domain Complexed with A Quinazoline Inhibitor- GW572016, PDB code: 1xkk:
Chlorine binding site 1 out of 1 in 1xkk
Go back to
Chlorine binding site 1 out
of 1 in the Egfr Kinase Domain Complexed with A Quinazoline Inhibitor- GW572016
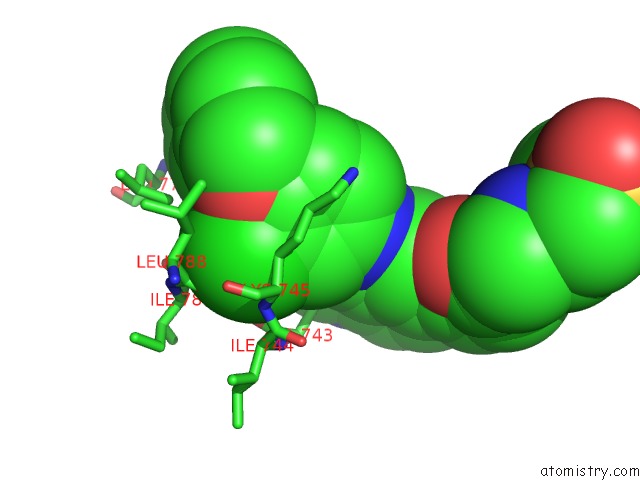
Mono view
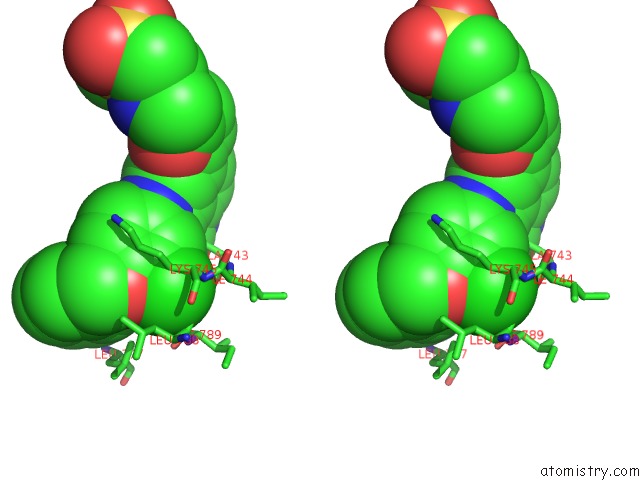
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Egfr Kinase Domain Complexed with A Quinazoline Inhibitor- GW572016 within 5.0Å range:
|
Reference:
E.R.Wood,
A.T.Truesdale,
O.B.Mcdonald,
D.Yuan,
A.Hassell,
S.H.Dickerson,
B.Ellis,
C.Pennisi,
E.Horne,
K.Lackey,
K.J.Alligood,
D.W.Rusnak,
T.M.Gilmer,
L.Shewchuk.
A Unique Structure For Epidermal Growth Factor Receptor Bound to GW572016 (Lapatinib): Relationships Among Protein Conformation, Inhibitor Off-Rate, and Receptor Activity in Tumor Cells. Cancer Res. V. 64 6652 2004.
ISSN: ISSN 0008-5472
PubMed: 15374980
DOI: 10.1158/0008-5472.CAN-04-1168
Page generated: Thu Jul 10 20:26:39 2025
ISSN: ISSN 0008-5472
PubMed: 15374980
DOI: 10.1158/0008-5472.CAN-04-1168
Last articles
Mg in 3BSUMg in 3BS1
Mg in 3BTX
Mg in 3BRW
Mg in 3BPD
Mg in 3BQB
Mg in 3BRE
Mg in 3BRB
Mg in 3BQP
Mg in 3BO3