Chlorine »
PDB 2fs7-2g7z »
2fxs »
Chlorine in PDB 2fxs: Yeast HSP82 in Complex with the Novel HSP90 Inhibitor Radamide
Protein crystallography data
The structure of Yeast HSP82 in Complex with the Novel HSP90 Inhibitor Radamide, PDB code: 2fxs
was solved by
R.M.Immormino,
D.T.Gewirth,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 44.41 / 2.00 |
| Space group | P 43 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 74.205, 74.205, 110.852, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19.6 / 22.6 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Yeast HSP82 in Complex with the Novel HSP90 Inhibitor Radamide
(pdb code 2fxs). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Yeast HSP82 in Complex with the Novel HSP90 Inhibitor Radamide, PDB code: 2fxs:
In total only one binding site of Chlorine was determined in the Yeast HSP82 in Complex with the Novel HSP90 Inhibitor Radamide, PDB code: 2fxs:
Chlorine binding site 1 out of 1 in 2fxs
Go back to
Chlorine binding site 1 out
of 1 in the Yeast HSP82 in Complex with the Novel HSP90 Inhibitor Radamide
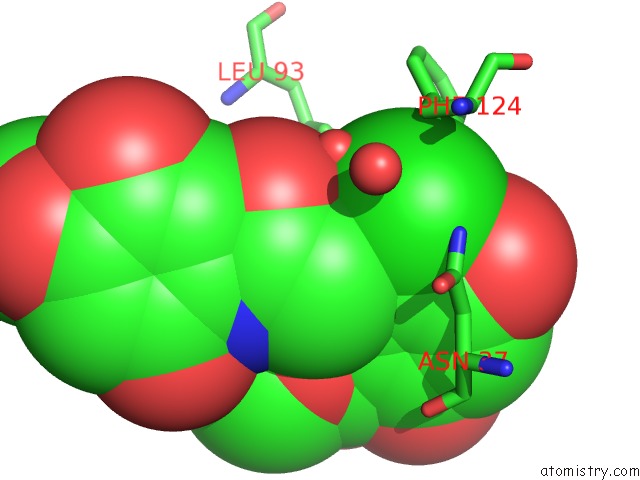
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Yeast HSP82 in Complex with the Novel HSP90 Inhibitor Radamide within 5.0Å range:
|
Reference:
R.M.Immormino,
L.E.Metzger,
P.N.Reardon,
D.E.Dollins,
B.S.Blagg,
D.T.Gewirth.
Different Poses For Ligand and Chaperone in Inhibitor-Bound HSP90 and GRP94: Implications For Paralog-Specific Drug Design. J.Mol.Biol. V. 388 1033 2009.
ISSN: ISSN 0022-2836
PubMed: 19361515
DOI: 10.1016/J.JMB.2009.03.071
Page generated: Thu Jul 10 22:13:31 2025
ISSN: ISSN 0022-2836
PubMed: 19361515
DOI: 10.1016/J.JMB.2009.03.071
Last articles
Fe in 5TISFe in 5TWT
Fe in 5TQN
Fe in 5TQO
Fe in 5TQP
Fe in 5TPG
Fe in 5TK5
Fe in 5TL8
Fe in 5T5I
Fe in 5TH5