Chlorine »
PDB 2gm9-2h9f »
2gvk »
Chlorine in PDB 2gvk: Crystal Structure of A Dye-Decolorizing Peroxidase (Dyp) From Bacteroides Thetaiotaomicron Vpi-5482 at 1.6 A Resolution
Protein crystallography data
The structure of Crystal Structure of A Dye-Decolorizing Peroxidase (Dyp) From Bacteroides Thetaiotaomicron Vpi-5482 at 1.6 A Resolution, PDB code: 2gvk
was solved by
Joint Center For Structural Genomics (Jcsg),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 28.01 / 1.60 |
| Space group | P 63 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 109.090, 109.090, 122.100, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 16.4 / 19 |
Other elements in 2gvk:
The structure of Crystal Structure of A Dye-Decolorizing Peroxidase (Dyp) From Bacteroides Thetaiotaomicron Vpi-5482 at 1.6 A Resolution also contains other interesting chemical elements:
| Sodium | (Na) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of A Dye-Decolorizing Peroxidase (Dyp) From Bacteroides Thetaiotaomicron Vpi-5482 at 1.6 A Resolution
(pdb code 2gvk). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 3 binding sites of Chlorine where determined in the Crystal Structure of A Dye-Decolorizing Peroxidase (Dyp) From Bacteroides Thetaiotaomicron Vpi-5482 at 1.6 A Resolution, PDB code: 2gvk:
Jump to Chlorine binding site number: 1; 2; 3;
In total 3 binding sites of Chlorine where determined in the Crystal Structure of A Dye-Decolorizing Peroxidase (Dyp) From Bacteroides Thetaiotaomicron Vpi-5482 at 1.6 A Resolution, PDB code: 2gvk:
Jump to Chlorine binding site number: 1; 2; 3;
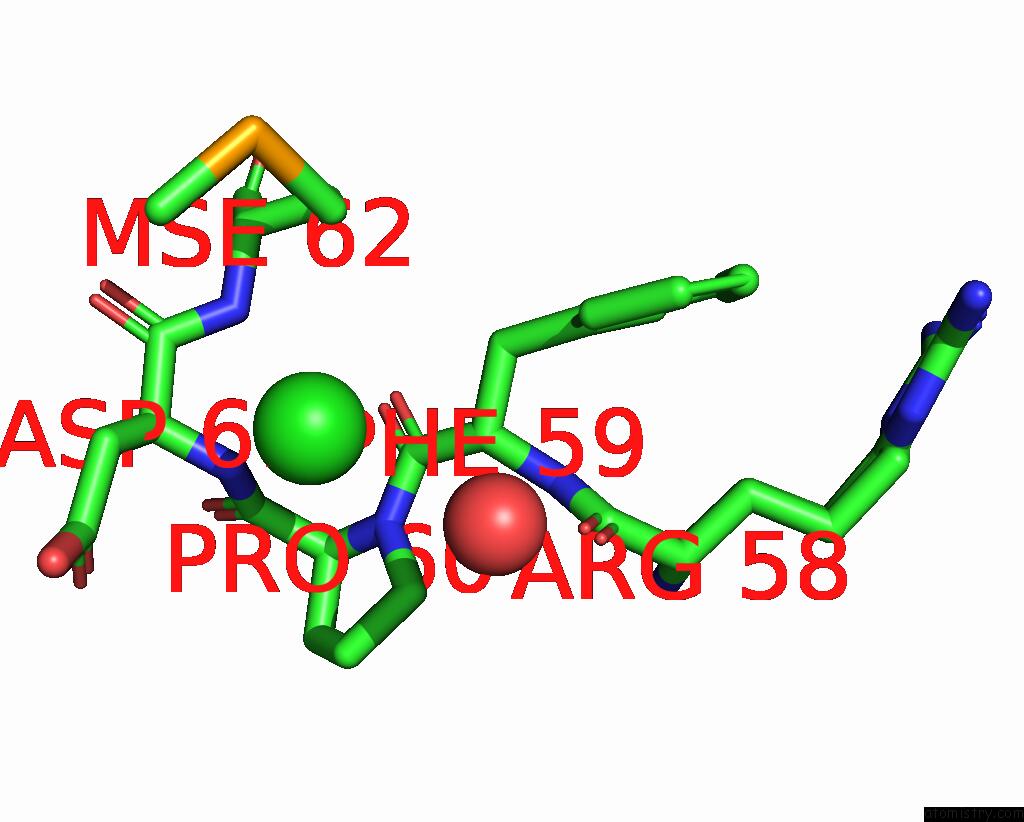
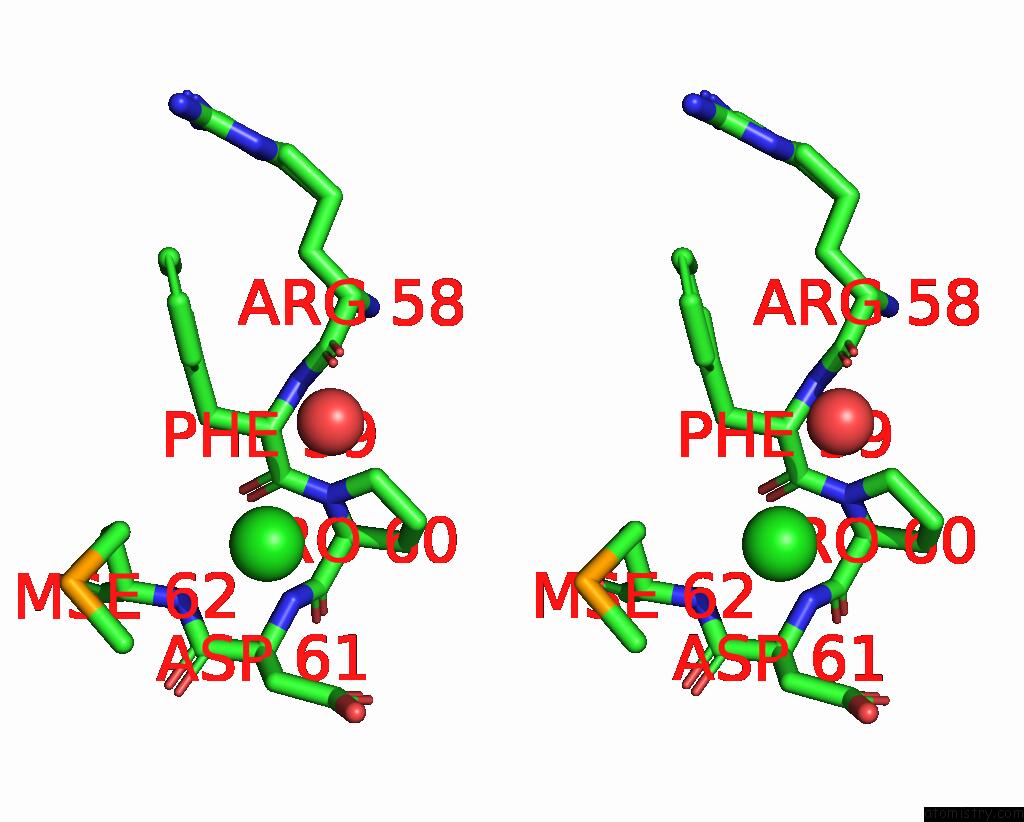
Chlorine binding site 1 out of 3 in 2gvk
Go back to
Chlorine binding site 1 out
of 3 in the Crystal Structure of A Dye-Decolorizing Peroxidase (Dyp) From Bacteroides Thetaiotaomicron Vpi-5482 at 1.6 A Resolution
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of A Dye-Decolorizing Peroxidase (Dyp) From Bacteroides Thetaiotaomicron Vpi-5482 at 1.6 A Resolution within 5.0Å range:
|
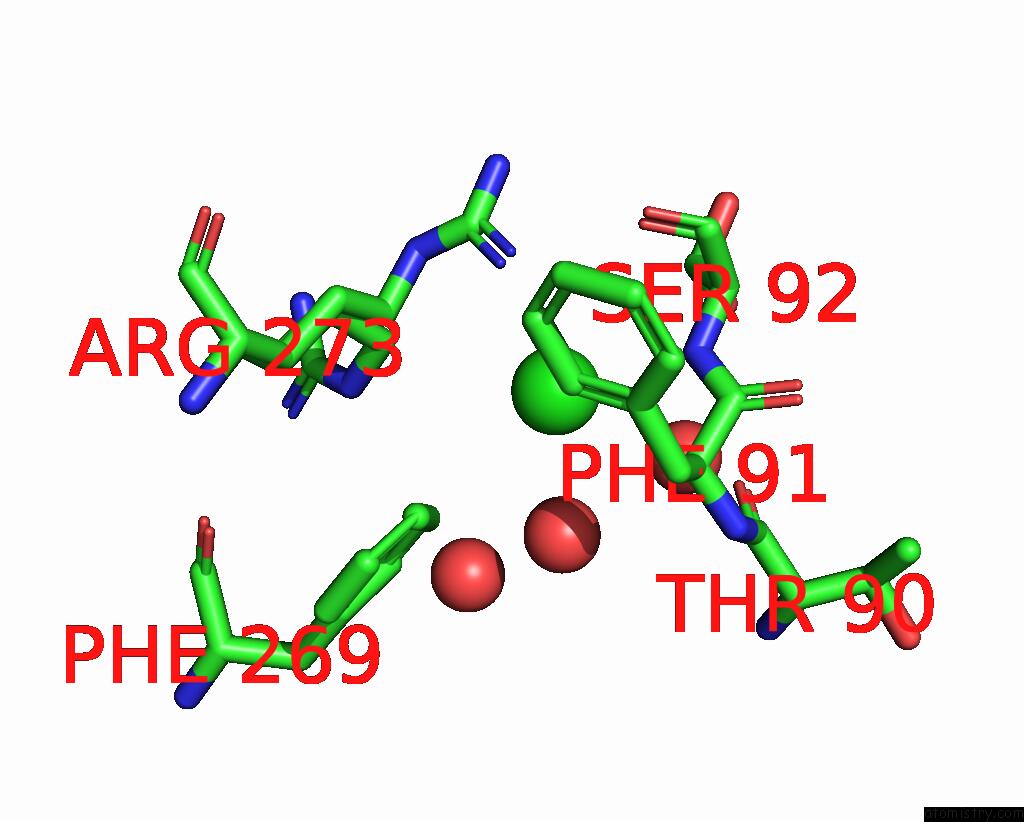
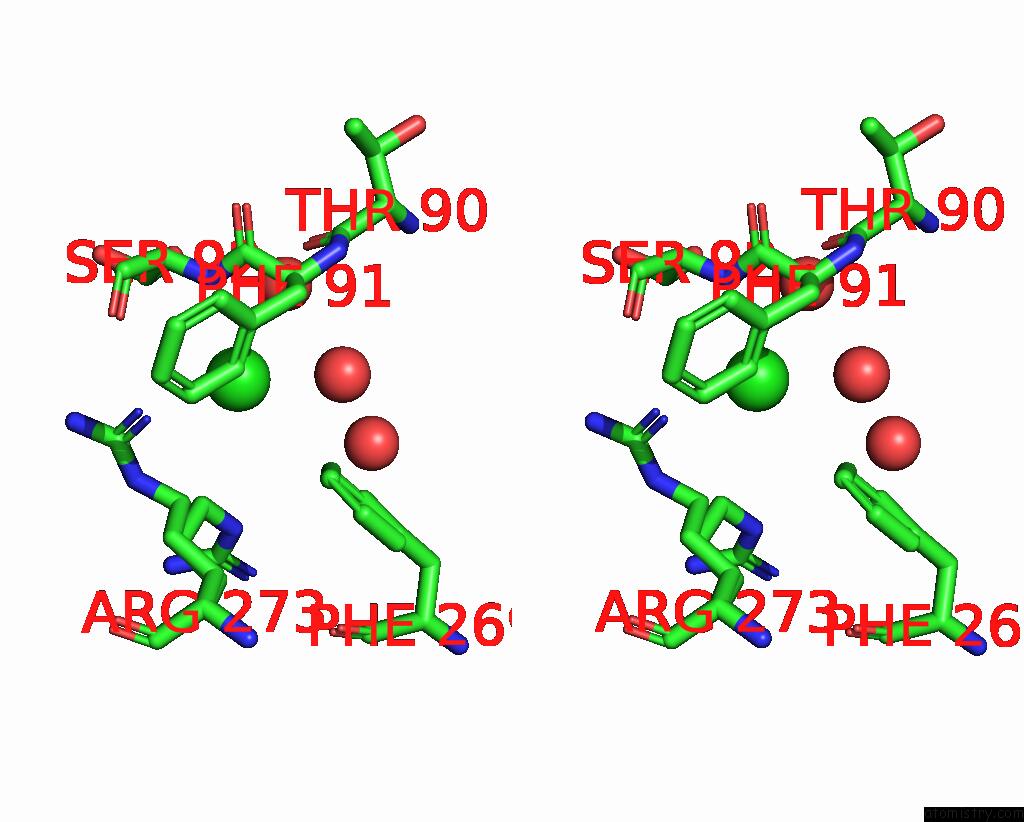
Chlorine binding site 2 out of 3 in 2gvk
Go back to
Chlorine binding site 2 out
of 3 in the Crystal Structure of A Dye-Decolorizing Peroxidase (Dyp) From Bacteroides Thetaiotaomicron Vpi-5482 at 1.6 A Resolution
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of A Dye-Decolorizing Peroxidase (Dyp) From Bacteroides Thetaiotaomicron Vpi-5482 at 1.6 A Resolution within 5.0Å range:
|
Chlorine binding site 3 out of 3 in 2gvk
Go back to
Chlorine binding site 3 out
of 3 in the Crystal Structure of A Dye-Decolorizing Peroxidase (Dyp) From Bacteroides Thetaiotaomicron Vpi-5482 at 1.6 A Resolution
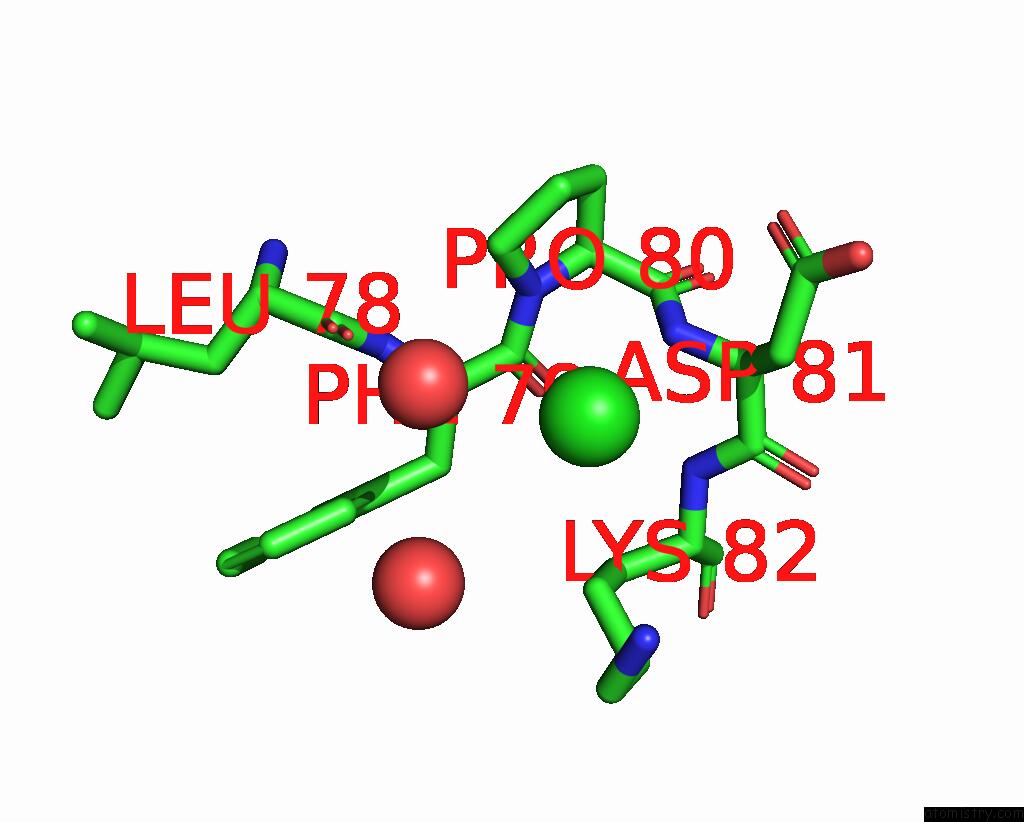
Mono view
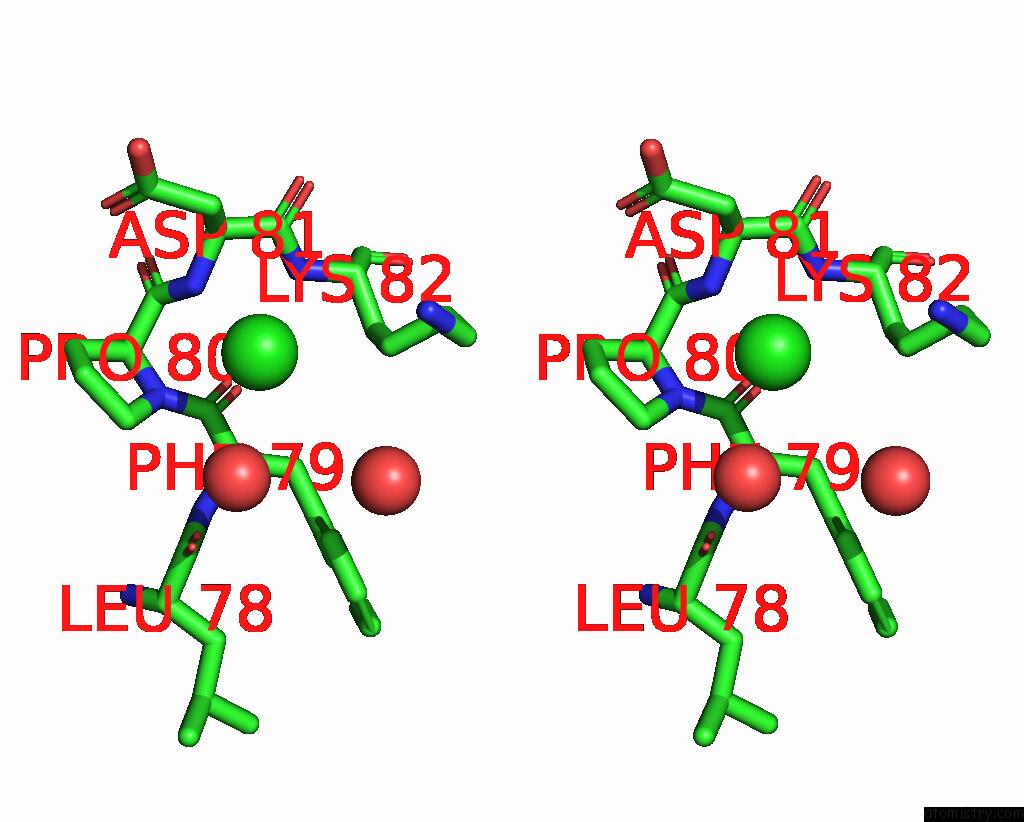
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of A Dye-Decolorizing Peroxidase (Dyp) From Bacteroides Thetaiotaomicron Vpi-5482 at 1.6 A Resolution within 5.0Å range:
|
Reference:
C.Zubieta,
S.S.Krishna,
M.Kapoor,
P.Kozbial,
D.Mcmullan,
H.L.Axelrod,
M.D.Miller,
P.Abdubek,
E.Ambing,
T.Astakhova,
D.Carlton,
H.J.Chiu,
T.Clayton,
M.C.Deller,
L.Duan,
M.A.Elsliger,
J.Feuerhelm,
S.K.Grzechnik,
J.Hale,
E.Hampton,
G.W.Han,
L.Jaroszewski,
K.K.Jin,
H.E.Klock,
M.W.Knuth,
A.Kumar,
D.Marciano,
A.T.Morse,
E.Nigoghossian,
L.Okach,
S.Oommachen,
R.Reyes,
C.L.Rife,
P.Schimmel,
H.Van Den Bedem,
D.Weekes,
A.White,
Q.Xu,
K.O.Hodgson,
J.Wooley,
A.M.Deacon,
A.Godzik,
S.A.Lesley,
I.A.Wilson.
Crystal Structures of Two Novel Dye-Decolorizing Peroxidases Reveal A Beta-Barrel Fold with A Conserved Heme-Binding Motif. Proteins V. 69 223 2007.
ISSN: ISSN 0887-3585
PubMed: 17654545
DOI: 10.1002/PROT.21550
Page generated: Thu Jul 10 22:27:59 2025
ISSN: ISSN 0887-3585
PubMed: 17654545
DOI: 10.1002/PROT.21550
Last articles
K in 4BYGK in 4BVA
K in 4BGA
K in 4BV8
K in 4BKX
K in 4BGB
K in 4BI3
K in 4BH5
K in 4B3T
K in 4BG8