Chlorine »
PDB 2gm9-2h9f »
2gz7 »
Chlorine in PDB 2gz7: Structure-Based Drug Design and Structural Biology Study of Novel Nonpeptide Inhibitors of Sars-Cov Main Protease
Protein crystallography data
The structure of Structure-Based Drug Design and Structural Biology Study of Novel Nonpeptide Inhibitors of Sars-Cov Main Protease, PDB code: 2gz7
was solved by
I.L.Lu,
S.Y.Wu,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.00 / 1.86 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 107.776, 82.777, 53.579, 90.00, 104.93, 90.00 |
| R / Rfree (%) | 20.4 / 23.3 |
Other elements in 2gz7:
The structure of Structure-Based Drug Design and Structural Biology Study of Novel Nonpeptide Inhibitors of Sars-Cov Main Protease also contains other interesting chemical elements:
| Fluorine | (F) | 3 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Structure-Based Drug Design and Structural Biology Study of Novel Nonpeptide Inhibitors of Sars-Cov Main Protease
(pdb code 2gz7). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Structure-Based Drug Design and Structural Biology Study of Novel Nonpeptide Inhibitors of Sars-Cov Main Protease, PDB code: 2gz7:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Structure-Based Drug Design and Structural Biology Study of Novel Nonpeptide Inhibitors of Sars-Cov Main Protease, PDB code: 2gz7:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 2gz7
Go back to
Chlorine binding site 1 out
of 2 in the Structure-Based Drug Design and Structural Biology Study of Novel Nonpeptide Inhibitors of Sars-Cov Main Protease
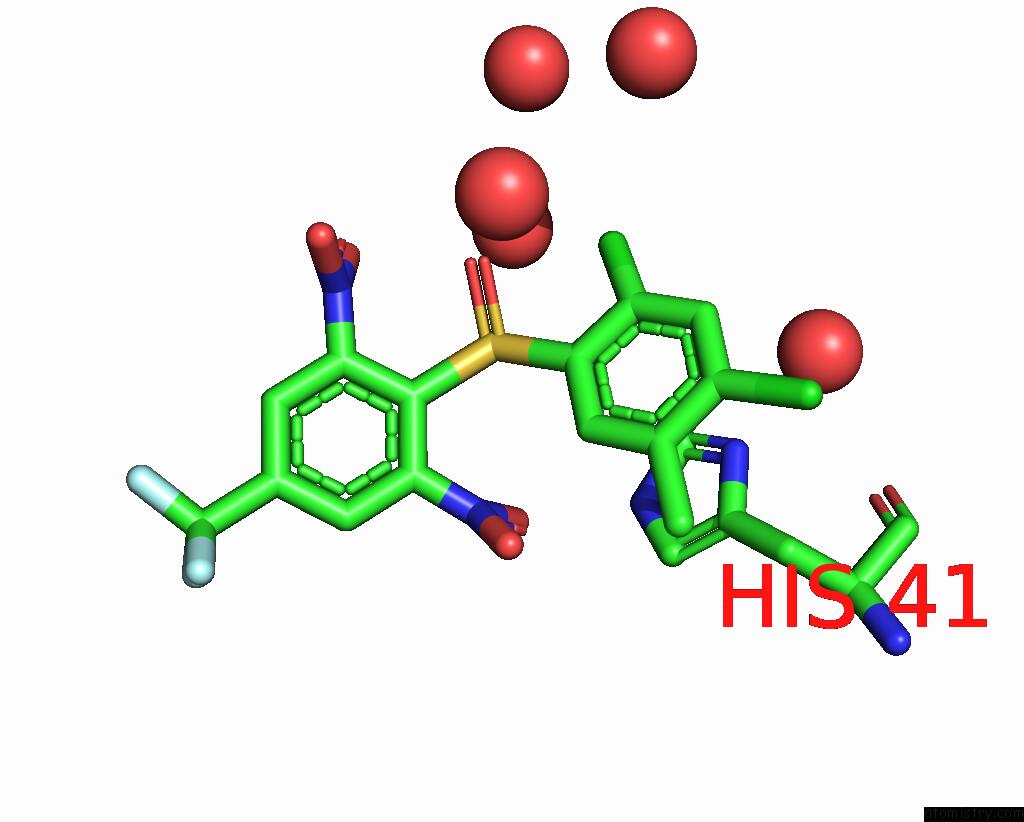
Mono view
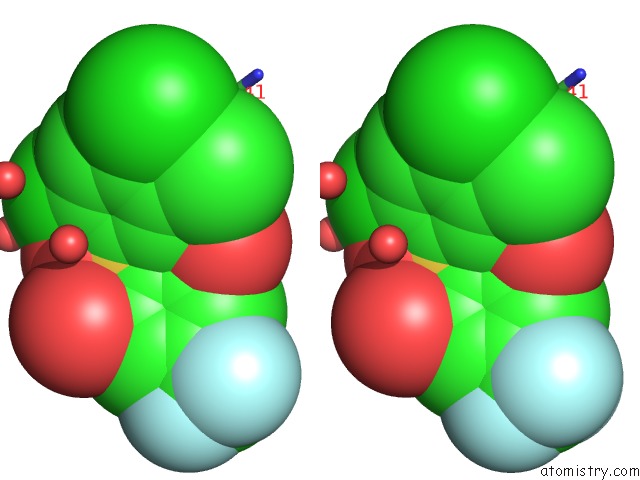
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Structure-Based Drug Design and Structural Biology Study of Novel Nonpeptide Inhibitors of Sars-Cov Main Protease within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 2gz7
Go back to
Chlorine binding site 2 out
of 2 in the Structure-Based Drug Design and Structural Biology Study of Novel Nonpeptide Inhibitors of Sars-Cov Main Protease
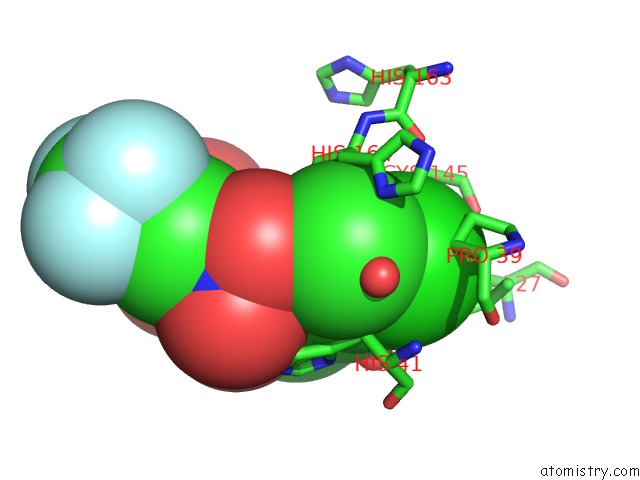
Mono view
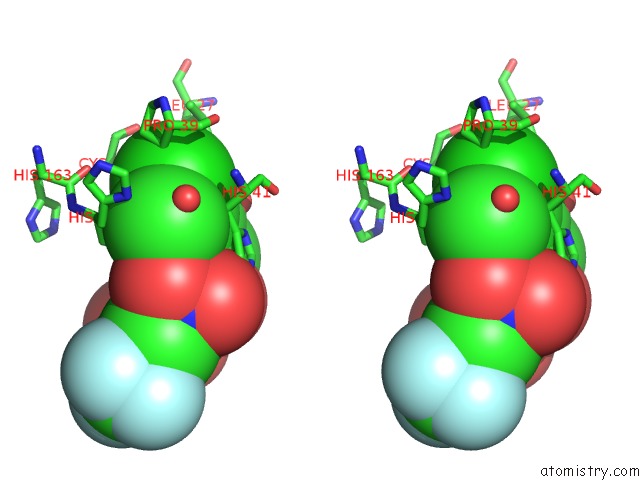
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Structure-Based Drug Design and Structural Biology Study of Novel Nonpeptide Inhibitors of Sars-Cov Main Protease within 5.0Å range:
|
Reference:
I.L.Lu,
N.Mahindroo,
P.H.Liang,
Y.H.Peng,
C.J.Kuo,
K.C.Tsai,
H.P.Hsieh,
Y.S.Chao,
S.Y.Wu.
Structure-Based Drug Design and Structural Biology Study of Novel Nonpeptide Inhibitors of Severe Acute Respiratory Syndrome Coronavirus Main Protease J.Med.Chem. V. 49 5154 2006.
ISSN: ISSN 0022-2623
PubMed: 16913704
DOI: 10.1021/JM060207O
Page generated: Thu Jul 10 22:28:53 2025
ISSN: ISSN 0022-2623
PubMed: 16913704
DOI: 10.1021/JM060207O
Last articles
K in 5OPXK in 5PA3
K in 5S9L
K in 5P9D
K in 5P9C
K in 5P9A
K in 5OX7
K in 5OWO
K in 5OU5
K in 5OSN