Chlorine »
PDB 2jh7-2krd »
2jlx »
Chlorine in PDB 2jlx: Dengue Virus 4 NS3 Helicase in Complex with Ssrna and Adp- Vanadate
Enzymatic activity of Dengue Virus 4 NS3 Helicase in Complex with Ssrna and Adp- Vanadate
All present enzymatic activity of Dengue Virus 4 NS3 Helicase in Complex with Ssrna and Adp- Vanadate:
3.4.21.91;
3.4.21.91;
Protein crystallography data
The structure of Dengue Virus 4 NS3 Helicase in Complex with Ssrna and Adp- Vanadate, PDB code: 2jlx
was solved by
D.H.Luo,
T.Xu,
R.P.Watson,
D.S.Becker,
A.Sampath,
W.Jahnke,
S.S.Yeong,
C.H.Wang,
S.P.Lim,
S.G.Vasudevan,
J.Lescar,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.00 / 2.20 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 132.537, 105.207, 72.352, 90.00, 117.62, 90.00 |
| R / Rfree (%) | 19.565 / 24.506 |
Other elements in 2jlx:
The structure of Dengue Virus 4 NS3 Helicase in Complex with Ssrna and Adp- Vanadate also contains other interesting chemical elements:
| Manganese | (Mn) | 2 atoms |
| Vanadium | (V) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Dengue Virus 4 NS3 Helicase in Complex with Ssrna and Adp- Vanadate
(pdb code 2jlx). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Dengue Virus 4 NS3 Helicase in Complex with Ssrna and Adp- Vanadate, PDB code: 2jlx:
In total only one binding site of Chlorine was determined in the Dengue Virus 4 NS3 Helicase in Complex with Ssrna and Adp- Vanadate, PDB code: 2jlx:
Chlorine binding site 1 out of 1 in 2jlx
Go back to
Chlorine binding site 1 out
of 1 in the Dengue Virus 4 NS3 Helicase in Complex with Ssrna and Adp- Vanadate
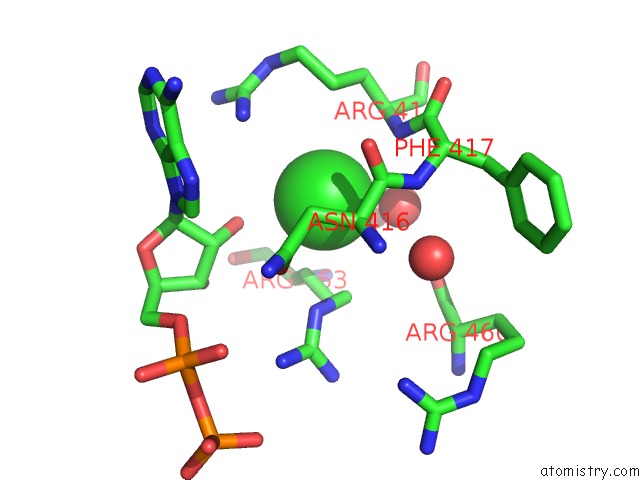
Mono view
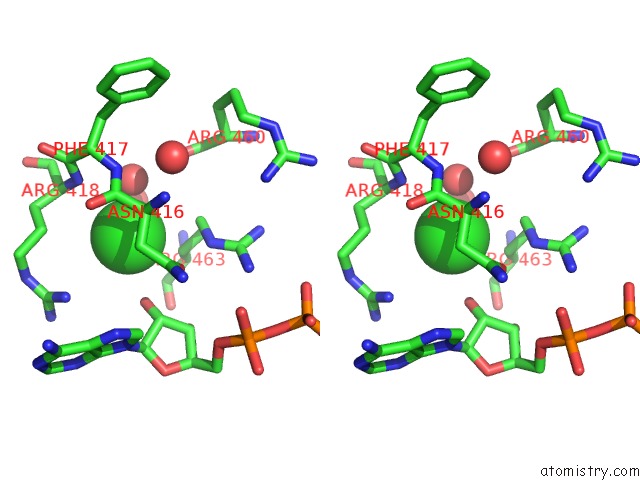
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Dengue Virus 4 NS3 Helicase in Complex with Ssrna and Adp- Vanadate within 5.0Å range:
|
Reference:
D.H.Luo,
T.Xu,
R.P.Watson,
D.Scherer-Becker,
A.Sampath,
W.Jahnke,
S.S.Yeong,
C.H.Wang,
S.P.Lim,
A.Strongin,
S.G.Vasudevan,
J.Lescar.
Insights Into Rna Unwinding and Atp Hydrolysis By the Flavivirus NS3 Protein. Embo J. V. 27 3209 2008.
ISSN: ISSN 0261-4189
PubMed: 19008861
DOI: 10.1038/EMBOJ.2008.232
Page generated: Thu Jul 10 23:15:31 2025
ISSN: ISSN 0261-4189
PubMed: 19008861
DOI: 10.1038/EMBOJ.2008.232
Last articles
Mg in 6QPHMg in 6QV0
Mg in 6QUZ
Mg in 6QUY
Mg in 6QUX
Mg in 6QUW
Mg in 6QUV
Mg in 6QUU
Mg in 6QUS
Mg in 6QTN