Chlorine »
PDB 2z0g-2zgd »
2z0l »
Chlorine in PDB 2z0l: Crystal Structure of Ebv-Dna Polymerase Accessory Protein BMRF1
Protein crystallography data
The structure of Crystal Structure of Ebv-Dna Polymerase Accessory Protein BMRF1, PDB code: 2z0l
was solved by
K.Murayama,
M.Kato-Murayama,
T.Terada,
M.Shirouzu,
S.Yokoyama,
Riken Structural Genomics/Proteomics Initiative (Rsgi),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.38 / 2.90 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 125.671, 191.635, 371.720, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.6 / 25.1 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Ebv-Dna Polymerase Accessory Protein BMRF1
(pdb code 2z0l). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 9 binding sites of Chlorine where determined in the Crystal Structure of Ebv-Dna Polymerase Accessory Protein BMRF1, PDB code: 2z0l:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9;
In total 9 binding sites of Chlorine where determined in the Crystal Structure of Ebv-Dna Polymerase Accessory Protein BMRF1, PDB code: 2z0l:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9;
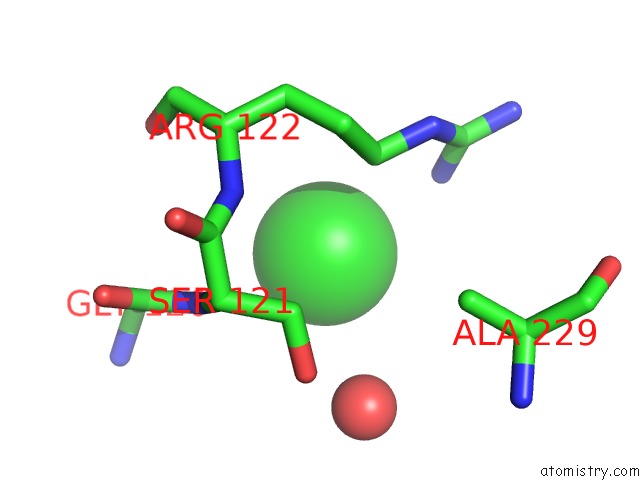
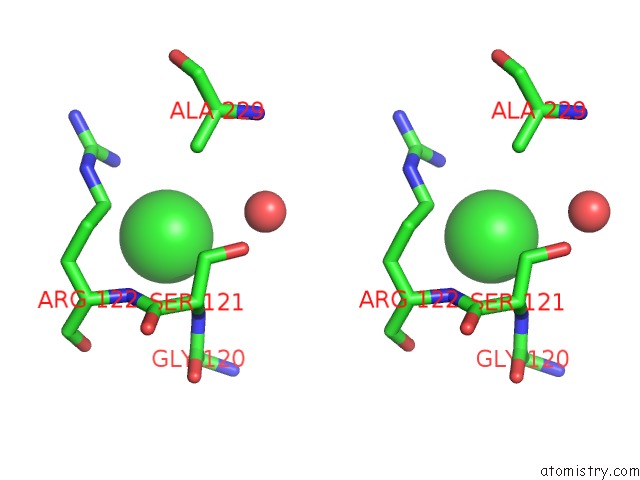
Chlorine binding site 1 out of 9 in 2z0l
Go back to
Chlorine binding site 1 out
of 9 in the Crystal Structure of Ebv-Dna Polymerase Accessory Protein BMRF1
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Ebv-Dna Polymerase Accessory Protein BMRF1 within 5.0Å range:
|
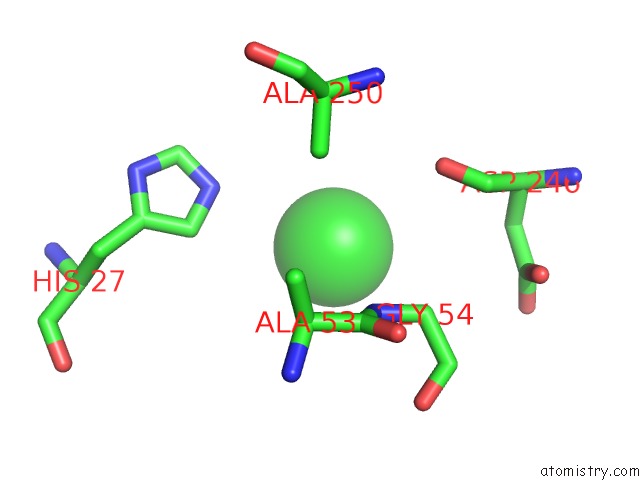
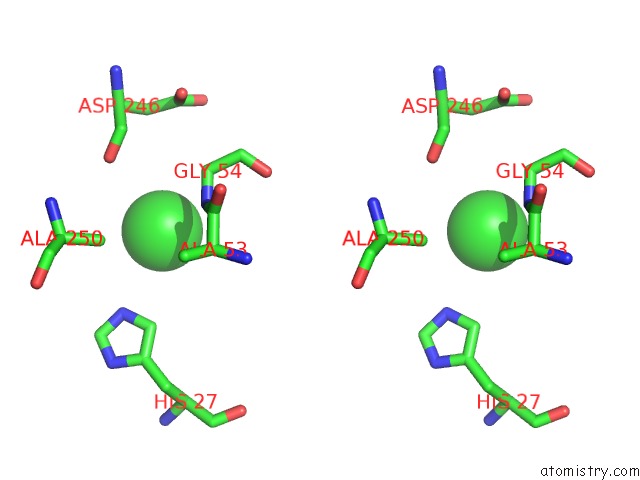
Chlorine binding site 2 out of 9 in 2z0l
Go back to
Chlorine binding site 2 out
of 9 in the Crystal Structure of Ebv-Dna Polymerase Accessory Protein BMRF1
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Ebv-Dna Polymerase Accessory Protein BMRF1 within 5.0Å range:
|
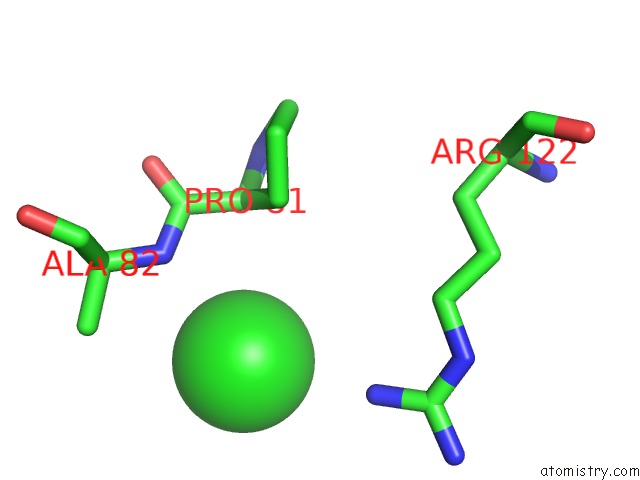
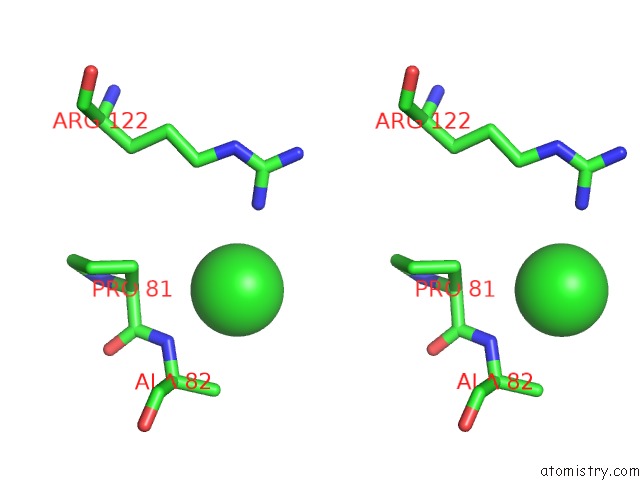
Chlorine binding site 3 out of 9 in 2z0l
Go back to
Chlorine binding site 3 out
of 9 in the Crystal Structure of Ebv-Dna Polymerase Accessory Protein BMRF1
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of Ebv-Dna Polymerase Accessory Protein BMRF1 within 5.0Å range:
|
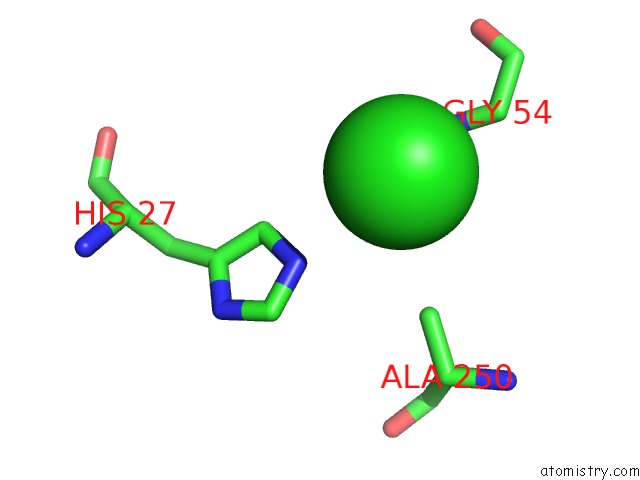
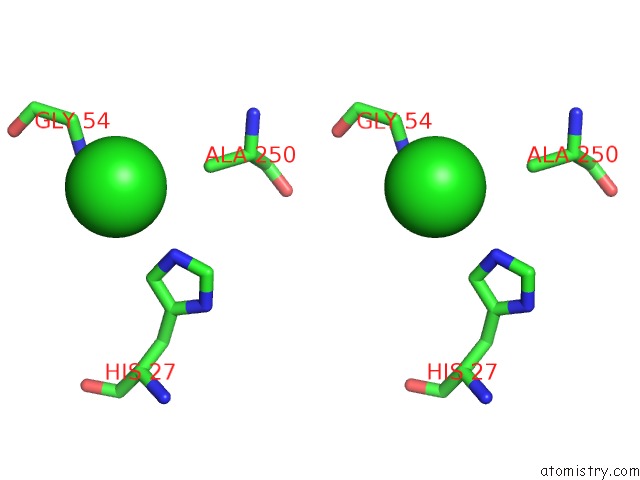
Chlorine binding site 4 out of 9 in 2z0l
Go back to
Chlorine binding site 4 out
of 9 in the Crystal Structure of Ebv-Dna Polymerase Accessory Protein BMRF1
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of Ebv-Dna Polymerase Accessory Protein BMRF1 within 5.0Å range:
|
Chlorine binding site 5 out of 9 in 2z0l
Go back to
Chlorine binding site 5 out
of 9 in the Crystal Structure of Ebv-Dna Polymerase Accessory Protein BMRF1
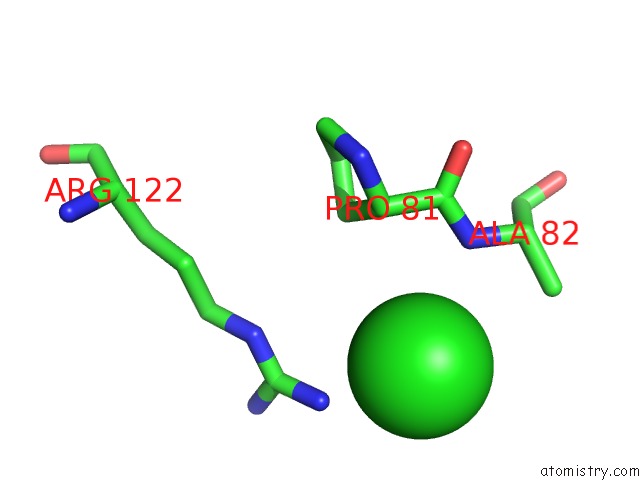
Mono view
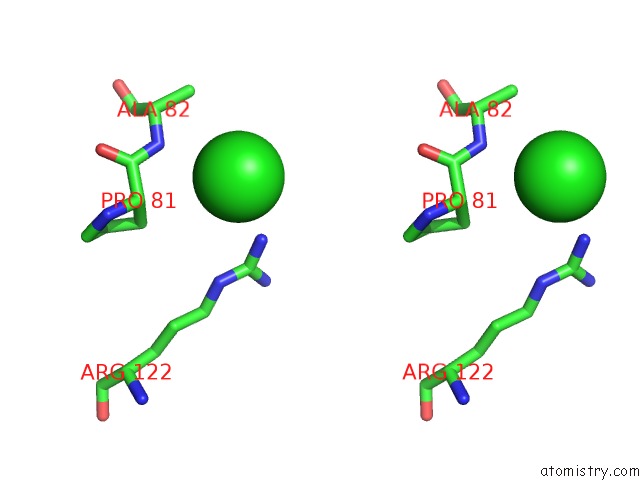
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Crystal Structure of Ebv-Dna Polymerase Accessory Protein BMRF1 within 5.0Å range:
|
Chlorine binding site 6 out of 9 in 2z0l
Go back to
Chlorine binding site 6 out
of 9 in the Crystal Structure of Ebv-Dna Polymerase Accessory Protein BMRF1
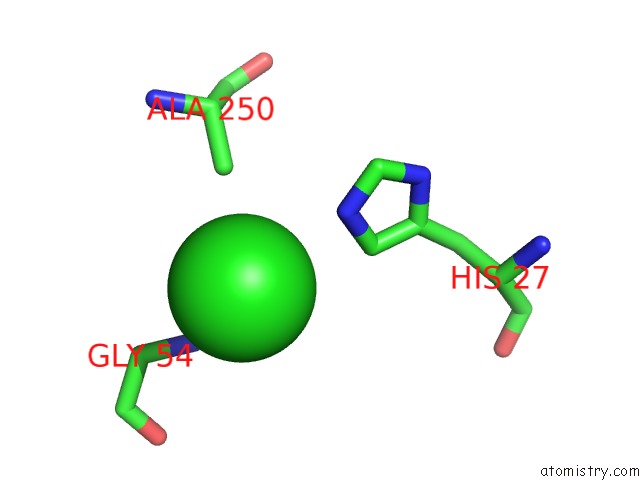
Mono view
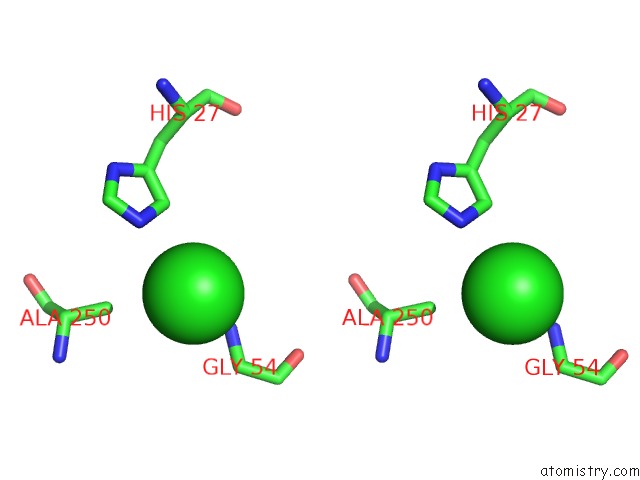
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Crystal Structure of Ebv-Dna Polymerase Accessory Protein BMRF1 within 5.0Å range:
|
Chlorine binding site 7 out of 9 in 2z0l
Go back to
Chlorine binding site 7 out
of 9 in the Crystal Structure of Ebv-Dna Polymerase Accessory Protein BMRF1
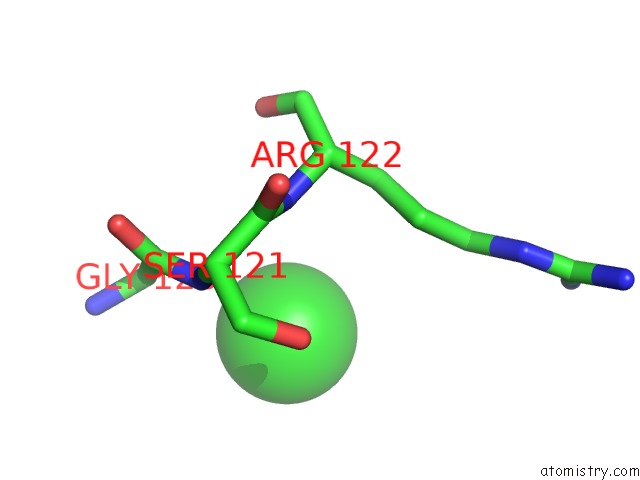
Mono view
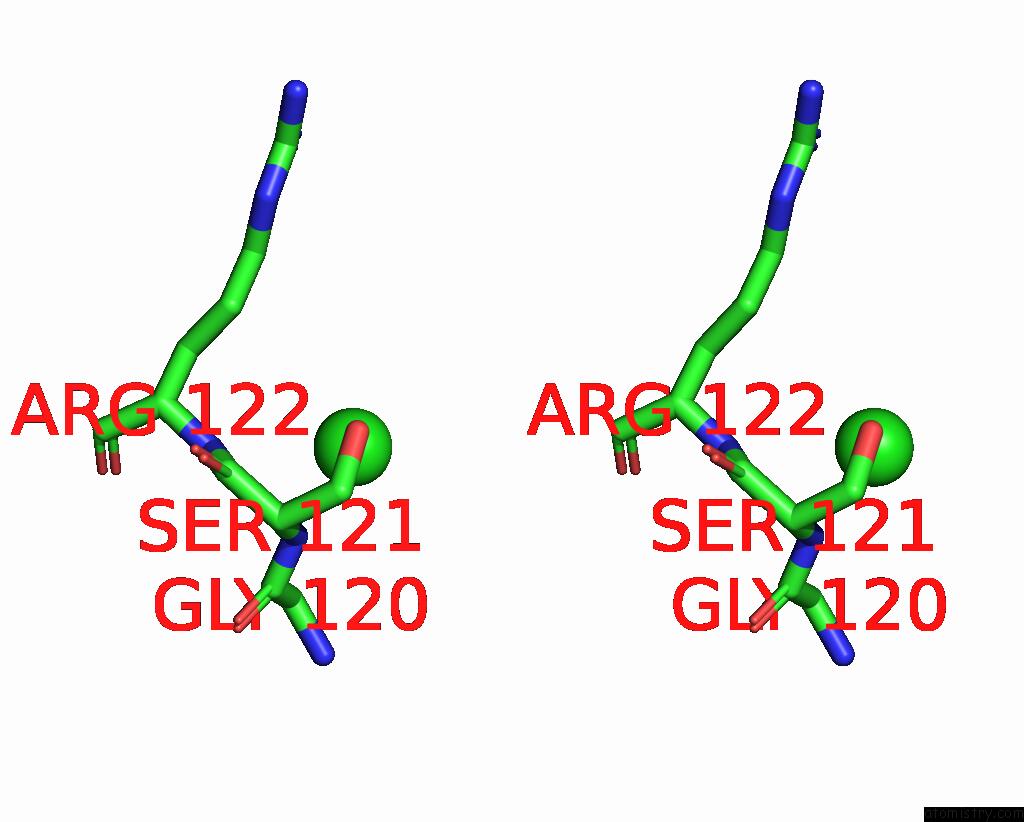
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 7 of Crystal Structure of Ebv-Dna Polymerase Accessory Protein BMRF1 within 5.0Å range:
|
Chlorine binding site 8 out of 9 in 2z0l
Go back to
Chlorine binding site 8 out
of 9 in the Crystal Structure of Ebv-Dna Polymerase Accessory Protein BMRF1
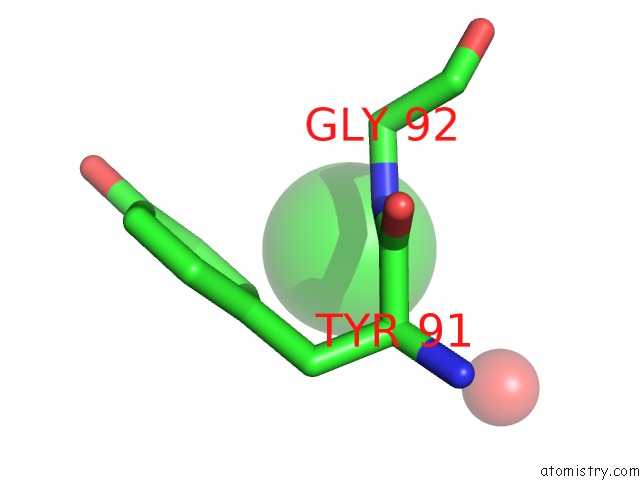
Mono view
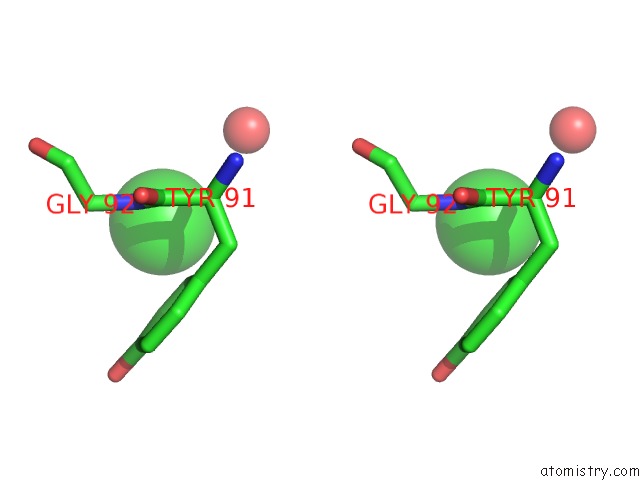
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 8 of Crystal Structure of Ebv-Dna Polymerase Accessory Protein BMRF1 within 5.0Å range:
|
Chlorine binding site 9 out of 9 in 2z0l
Go back to
Chlorine binding site 9 out
of 9 in the Crystal Structure of Ebv-Dna Polymerase Accessory Protein BMRF1
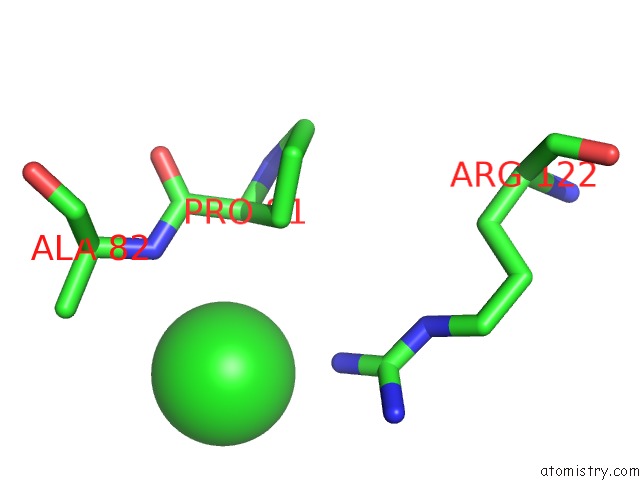
Mono view
Stereo pair view

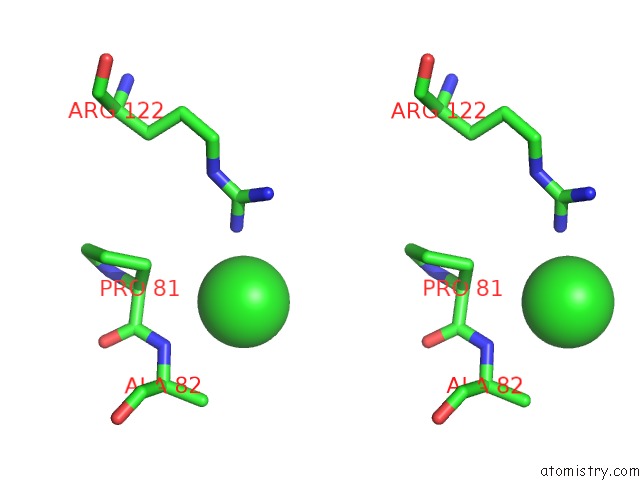
Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 9 of Crystal Structure of Ebv-Dna Polymerase Accessory Protein BMRF1 within 5.0Å range:
|
Reference:
K.Murayama,
S.Nakayama,
M.Kato-Murayama,
R.Akasaka,
N.Ohbayashi,
Y.Kamewari-Hayami,
T.Terada,
M.Shirouzu,
T.Tsurumi,
S.Yokoyama.
Crystal Structure of Epstein-Barr Virus Dna Polymerase Processivity Factor BMRF1 J.Biol.Chem. V. 284 35896 2009.
ISSN: ISSN 0021-9258
PubMed: 19801550
DOI: 10.1074/JBC.M109.051581
Page generated: Fri Jul 11 02:48:21 2025
ISSN: ISSN 0021-9258
PubMed: 19801550
DOI: 10.1074/JBC.M109.051581
Last articles
Mg in 5MMJMg in 5MRA
Mg in 5MTV
Mg in 5MS0
Mg in 5MRU
Mg in 5MQJ
Mg in 5MQW
Mg in 5MQT
Mg in 5MQL
Mg in 5MQ1