Chlorine »
PDB 2z0g-2zgd »
2z2d »
Chlorine in PDB 2z2d: Solution Structure of Human Macrophage Elastase (Mmp-12) Catalytic Domain Complexed with A Gamma-Keto Butanoic Acid Inhibitor
Enzymatic activity of Solution Structure of Human Macrophage Elastase (Mmp-12) Catalytic Domain Complexed with A Gamma-Keto Butanoic Acid Inhibitor
All present enzymatic activity of Solution Structure of Human Macrophage Elastase (Mmp-12) Catalytic Domain Complexed with A Gamma-Keto Butanoic Acid Inhibitor:
3.4.24.65;
3.4.24.65;
Other elements in 2z2d:
The structure of Solution Structure of Human Macrophage Elastase (Mmp-12) Catalytic Domain Complexed with A Gamma-Keto Butanoic Acid Inhibitor also contains other interesting chemical elements:
| Calcium | (Ca) | 45 atoms |
| Zinc | (Zn) | 30 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Solution Structure of Human Macrophage Elastase (Mmp-12) Catalytic Domain Complexed with A Gamma-Keto Butanoic Acid Inhibitor
(pdb code 2z2d). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Solution Structure of Human Macrophage Elastase (Mmp-12) Catalytic Domain Complexed with A Gamma-Keto Butanoic Acid Inhibitor, PDB code: 2z2d:
In total only one binding site of Chlorine was determined in the Solution Structure of Human Macrophage Elastase (Mmp-12) Catalytic Domain Complexed with A Gamma-Keto Butanoic Acid Inhibitor, PDB code: 2z2d:
Chlorine binding site 1 out of 1 in 2z2d
Go back to
Chlorine binding site 1 out
of 1 in the Solution Structure of Human Macrophage Elastase (Mmp-12) Catalytic Domain Complexed with A Gamma-Keto Butanoic Acid Inhibitor
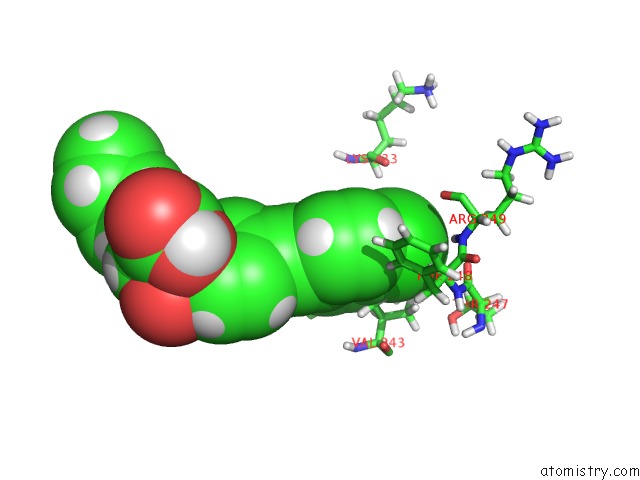
Mono view
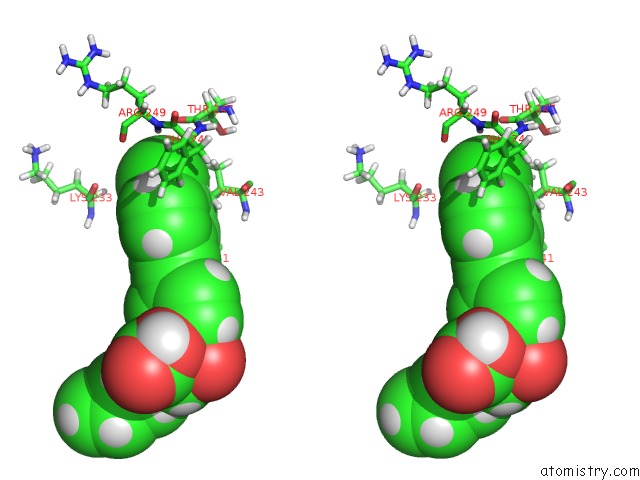
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Solution Structure of Human Macrophage Elastase (Mmp-12) Catalytic Domain Complexed with A Gamma-Keto Butanoic Acid Inhibitor within 5.0Å range:
|
Reference:
X.Zheng,
L.Ou.
Solution Structure of Human Macrophage Elastase (Mmp-12) Catalytic Domain Complexed with A Gamma-Keto Butanoic Acid Inhibitor To Be Published.
Page generated: Fri Jul 11 02:49:35 2025
Last articles
Mg in 364DMg in 3A0T
Mg in 357D
Mg in 3A06
Mg in 392D
Mg in 389D
Mg in 388D
Mg in 384D
Mg in 354D
Mg in 383D