Chlorine »
PDB 3bga-3bpr »
3bjz »
Chlorine in PDB 3bjz: Crystal Structure of Pseudomonas Aeruginosa Phosphoheptose Isomerase
Protein crystallography data
The structure of Crystal Structure of Pseudomonas Aeruginosa Phosphoheptose Isomerase, PDB code: 3bjz
was solved by
J.R.Walker,
E.Evdokimova,
M.Kudritska,
J.Osipiuk,
A.Joachimiak,
A.Savchenko,
Midwest Center For Structural Genomics (Mcsg),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 39.19 / 2.40 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 123.842, 131.609, 48.774, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19.7 / 27.4 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Pseudomonas Aeruginosa Phosphoheptose Isomerase
(pdb code 3bjz). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Crystal Structure of Pseudomonas Aeruginosa Phosphoheptose Isomerase, PDB code: 3bjz:
In total only one binding site of Chlorine was determined in the Crystal Structure of Pseudomonas Aeruginosa Phosphoheptose Isomerase, PDB code: 3bjz:
Chlorine binding site 1 out of 1 in 3bjz
Go back to
Chlorine binding site 1 out
of 1 in the Crystal Structure of Pseudomonas Aeruginosa Phosphoheptose Isomerase
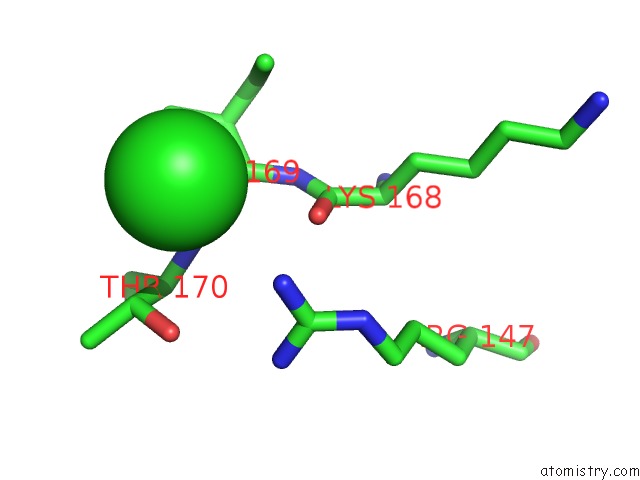
Mono view

Stereo pair view

Mono view

Stereo pair view
|
|
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Pseudomonas Aeruginosa Phosphoheptose Isomerase within 5.0Å range:
|
Reference:
P.L.Taylor,
K.M.Blakely,
G.P.De Leon,
J.R.Walker,
F.Mcarthur,
E.Evdokimova,
K.Zhang,
M.A.Valvano,
G.D.Wright,
M.S.Junop.
Structure and Function of Sedoheptulose-7-Phosphate Isomerase, A Critical Enzyme For Lipopolysaccharide Biosynthesis and A Target For Antibiotic Adjuvants. J.Biol.Chem. V. 283 2835 2008.
ISSN: ISSN 0021-9258
PubMed: 18056714
DOI: 10.1074/JBC.M706163200
Page generated: Fri Jul 11 03:28:44 2025
ISSN: ISSN 0021-9258
PubMed: 18056714
DOI: 10.1074/JBC.M706163200
Last articles
Mg in 4RRIMg in 4RRF
Mg in 4RRH
Mg in 4RRD
Mg in 4RRA
Mg in 4RR9
Mg in 4RR8
Mg in 4RR7
Mg in 4RQ5
Mg in 4RQI