Chlorine »
PDB 3ccq-3ck1 »
3cf8 »
Chlorine in PDB 3cf8: Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (Fabz) From Helicobacter Pylori in Complex with Quercetin
Protein crystallography data
The structure of Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (Fabz) From Helicobacter Pylori in Complex with Quercetin, PDB code: 3cf8
was solved by
L.Zhang,
D.Wu,
W.Liu,
X.Shen,
H.Jiang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.40 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 73.987, 100.438, 186.009, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19.3 / 22.6 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (Fabz) From Helicobacter Pylori in Complex with Quercetin
(pdb code 3cf8). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 6 binding sites of Chlorine where determined in the Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (Fabz) From Helicobacter Pylori in Complex with Quercetin, PDB code: 3cf8:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Chlorine where determined in the Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (Fabz) From Helicobacter Pylori in Complex with Quercetin, PDB code: 3cf8:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
Chlorine binding site 1 out of 6 in 3cf8
Go back to
Chlorine binding site 1 out
of 6 in the Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (Fabz) From Helicobacter Pylori in Complex with Quercetin
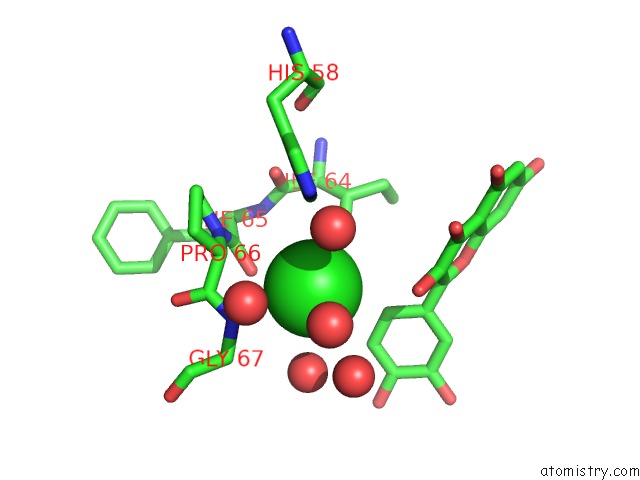
Mono view
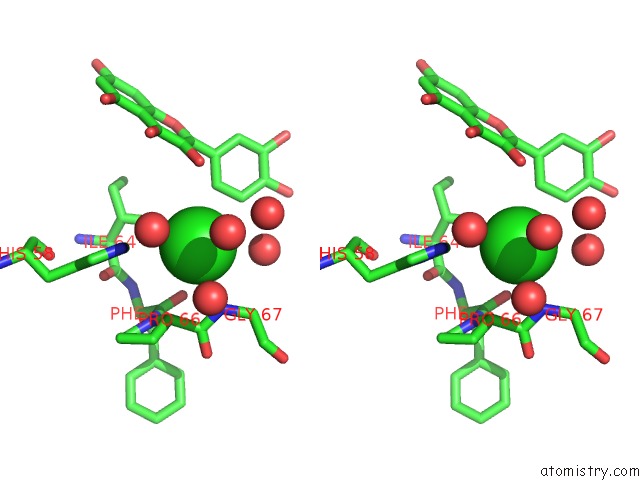
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (Fabz) From Helicobacter Pylori in Complex with Quercetin within 5.0Å range:
|
Chlorine binding site 2 out of 6 in 3cf8
Go back to
Chlorine binding site 2 out
of 6 in the Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (Fabz) From Helicobacter Pylori in Complex with Quercetin
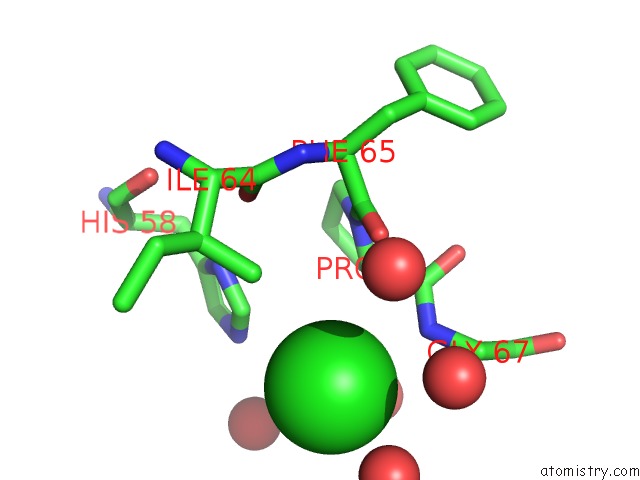
Mono view
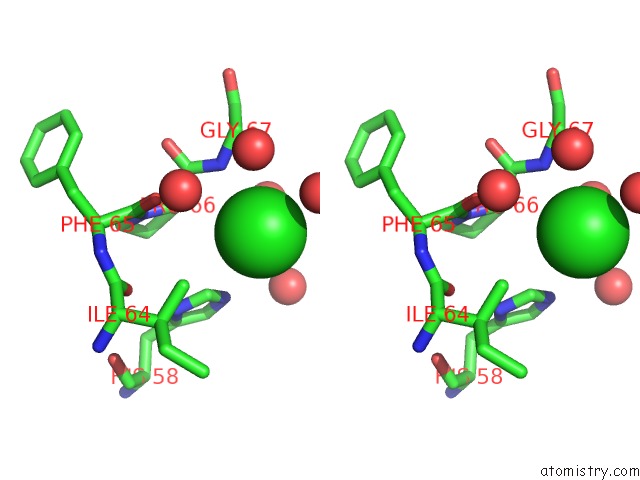
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (Fabz) From Helicobacter Pylori in Complex with Quercetin within 5.0Å range:
|
Chlorine binding site 3 out of 6 in 3cf8
Go back to
Chlorine binding site 3 out
of 6 in the Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (Fabz) From Helicobacter Pylori in Complex with Quercetin
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (Fabz) From Helicobacter Pylori in Complex with Quercetin within 5.0Å range:
|
Chlorine binding site 4 out of 6 in 3cf8
Go back to
Chlorine binding site 4 out
of 6 in the Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (Fabz) From Helicobacter Pylori in Complex with Quercetin
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (Fabz) From Helicobacter Pylori in Complex with Quercetin within 5.0Å range:
|
Chlorine binding site 5 out of 6 in 3cf8
Go back to
Chlorine binding site 5 out
of 6 in the Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (Fabz) From Helicobacter Pylori in Complex with Quercetin
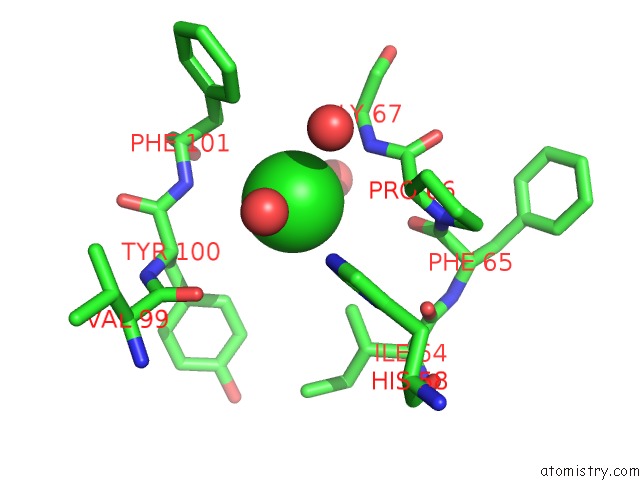
Mono view
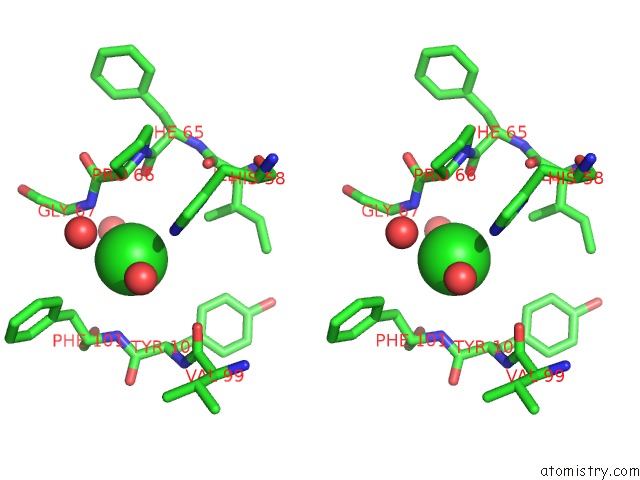
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (Fabz) From Helicobacter Pylori in Complex with Quercetin within 5.0Å range:
|
Chlorine binding site 6 out of 6 in 3cf8
Go back to
Chlorine binding site 6 out
of 6 in the Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (Fabz) From Helicobacter Pylori in Complex with Quercetin
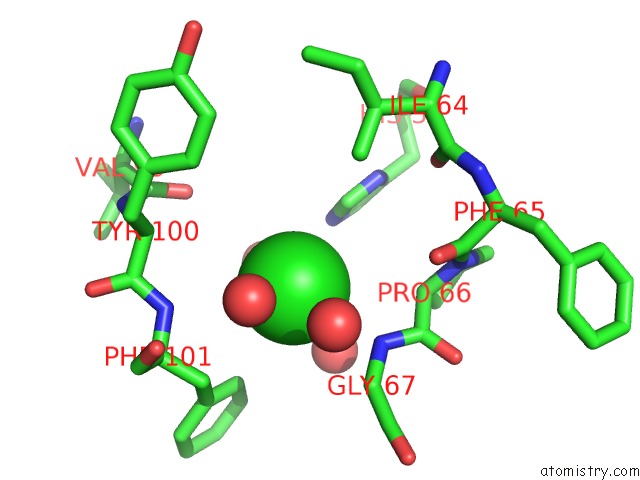
Mono view
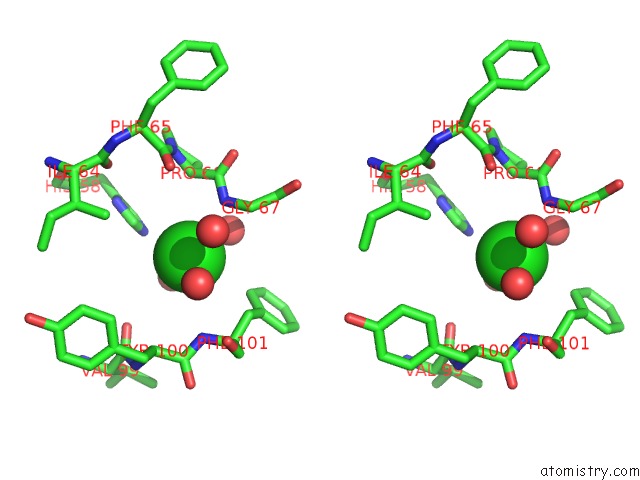
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Crystal Structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (Fabz) From Helicobacter Pylori in Complex with Quercetin within 5.0Å range:
|
Reference:
L.Zhang,
Y.Kong,
D.Wu,
H.Zhang,
J.Wu,
J.Chen,
J.Ding,
L.Hu,
H.Jiang,
X.Shen.
Three Flavonoids Targeting the Beta-Hydroxyacyl-Acyl Carrier Protein Dehydratase From Helicobacter Pylori: Crystal Structure Characterization with Enzymatic Inhibition Assay Protein Sci. V. 17 1971 2008.
ISSN: ISSN 0961-8368
PubMed: 18780820
DOI: 10.1110/PS.036186.108
Page generated: Fri Jul 11 04:00:22 2025
ISSN: ISSN 0961-8368
PubMed: 18780820
DOI: 10.1110/PS.036186.108
Last articles
K in 2FCXK in 2DCN
K in 2FBW
K in 2FCA
K in 2F1H
K in 2EZ2
K in 2DV6
K in 2EZ1
K in 2C9D
K in 2DWU